Next: Mixture of Bernoulli Up: Clustering Analysis Previous: K-means clustering
In K-means clustering, each sample point 

This issue can be addressed by the method of
Gaussian mixture model (GMM) based on the assumption
that each of the 




![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img9.svg)

 is the weight for the kth Gaussian
is the weight for the kth Gaussian
 , satisfying
, satisfying
 |
(210) |
 are estimated based on the given dataset, and then each sample in the
dataset is assigned to one of the clusters.
are estimated based on the given dataset, and then each sample in the
dataset is assigned to one of the clusters.
We note that the GMM model in Eq. (209) is actually the same
as Eq. (11) in the naive Bayes classification. These two
methods are similar in the sense that each cluster or classe 







![${\bf z}=[z_1,\cdots,z_K]^T$](img784.svg)









We further introduce the following probabilities for each of the


 in the dataset to belong to
in the dataset to belong to  , represented
by
, represented
by  :
These
:
These  prior probabilities add up to 1, i.e, the
prior probabilities add up to 1, i.e, the  events
events
 are mutually exclusive and complementary,
as any sample
are mutually exclusive and complementary,
as any sample  belongs to one and only one of the
belongs to one and only one of the  clusters.
clusters.
 , assumed
to be a Gaussian:
, assumed
to be a Gaussian:
 and
and  :
:
 , we get the Gaussian mixture model, the distribution
, we get the Gaussian mixture model, the distribution
 of any sample
of any sample  regardless to which cluster
it belongs:
Note that Eqs. (211) through (214) are the
same as Eqs. (8) through (11) in the naive
Bayes classifier, respectively.
regardless to which cluster
it belongs:
Note that Eqs. (211) through (214) are the
same as Eqs. (8) through (11) in the naive
Bayes classifier, respectively.
All such probabilities defined for 
![${\bf z}=[z_1,\cdots,z_K]^T$](img784.svg)

 |
 |
 |
(215) |
 |
 |
 |
(216) |
 |
 |
 |
(217) |
Given the dataset
![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img9.svg)


![${\bf Z}=[{\bf z}_1,\cdots,{\bf z}_N]$](img801.svg)
![${\bf z}_n=[z_{n1},\cdots,z_{nK}]^T$](img802.svg)






![${\bf Z}=[{\bf z}_1,\cdots,{\bf z}_N]$](img801.svg)
![${\bf Y}=[{\bf y}_1,\cdots,{\bf y}_N]$](img806.svg)



 |
(218) |
 to
be estimated can be expressed as:
to
be estimated can be expressed as:
 as those that maximize
the likelihood function
as those that maximize
the likelihood function
 or its log function
or its log function
 , here we find the model parameters in
, here we find the model parameters in
 as those
that maximize the expectation of the log likelihood function above
with respect to the latent variables in
as those
that maximize the expectation of the log likelihood function above
with respect to the latent variables in  in the following
two iterative steps of the EM method:
in the following
two iterative steps of the EM method:
We first find the posterior probability for an observed 




 has to belong to one of the
has to belong to one of the  clusters, these
clusters, these
 posterior probabilities add up to 1, same ss the prior
probabilities
posterior probabilities add up to 1, same ss the prior
probabilities
 :
:
 |
(222) |
 can be considered as the responsibility cluster
can be considered as the responsibility cluster  takes
for
takes
for  . Once we have available all parameters on the
right-hand side of Eq. (221) for
. Once we have available all parameters on the
right-hand side of Eq. (221) for  , it is used
to assign each sample
, it is used
to assign each sample  to a cluster
to a cluster  with the
maximun responsibility
with the
maximun responsibility
 .
.
We note that the definition of 

We also note that the posterior probability 









We can now find the expectation of the log likelihood with respect
to the latent variables in 
 |
(224) |
We first set to zero the derivatives of the expectation of
the log likelihood with respect to each of the parameters in

 :
:
As all 


![$\displaystyle L(\theta,\,\lambda)
=\sum_{n=1}^N \sum_{k=1}^K P_{nk}
\left[\log ...
...bf x}_n,{\bf m}_k,{\bf\Sigma}_k)\right]
+\lambda\left(\sum_{k=1}^K P_k-1\right)$](img836.svg) |
(225) |
 to zero:
to zero:
 |
 |
![$\displaystyle \frac{\partial}{\partial P_k} \left[
\sum_{n=1}^N \sum_{k=1}^K P_...
...{\bf m}_k,{\bf\Sigma}_k)\right]
+\lambda\left(\sum_{k=1}^K P_k-1\right) \right]$](img838.svg) |
|
 |
 |
(226) |
 , we get
where we have defined
that satisfies
, we get
where we have defined
that satisfies
 |
(229) |
 , we get
, we get
 |
(230) |
 back into Eq. (227),
we get the expression for the prior
This is actually the same as the prior
back into Eq. (227),
we get the expression for the prior
This is actually the same as the prior  in Eq. (8)
in naive Bayes classification, but here
in Eq. (8)
in naive Bayes classification, but here  defined in
Eq. (228) is the sum of the probabilities for all
defined in
Eq. (228) is the sum of the probabilities for all  data samples to belong to
data samples to belong to  , instead of the number of data
samples in
, instead of the number of data
samples in  (unknown in this unsupervised case).
(unknown in this unsupervised case).
 :
:
 indicates that we have neglected
the scaling coefficient
indicates that we have neglected
the scaling coefficient
 of
the Gaussian, which is independent of
of
the Gaussian, which is independent of  . Multiplying
. Multiplying
 on both sides, we get
on both sides, we get
 |
(233) |
 we get
we get
 :
:
 |
(236) |
 on both sides of the equation above
we get
on both sides of the equation above
we get
 |
(237) |
 , we get
, we get
 found
respectively in Eqs. (231), (234), and
(238) in the M-step depend on
found
respectively in Eqs. (231), (234), and
(238) in the M-step depend on  in Eq. (221)
in the E-step, which in turn is a function of these parameters, i.e.,
the E-step and M-step need to be carried out in an alternative and
iterative fashion from some initial values of the parameters until
convergence.
in Eq. (221)
in the E-step, which in turn is a function of these parameters, i.e.,
the E-step and M-step need to be carried out in an alternative and
iterative fashion from some initial values of the parameters until
convergence.
In summary, here is the EM clustering algorithm based on Gaussian mixture model:
 , covariance
, covariance
 and
coefficient
and
coefficient  .
.
Find the responsibility 



 |
(239) |
Recalculate the parameters that maximize the likelihood function:
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
(240) |
 to belong to cluster
to belong to cluster
 is
is
 , and it is therefore
assigned to
, and it is therefore
assigned to  if
if
 .
.
We can show that the K-means algorithm is actually a special case
of the EM algorithm, when all covariance matrices are the same


 |
(241) |
 to belong to cluster
to belong to cluster
 is:
is:
 |
(242) |
 , all terms in the denominator approach
to zero, but the one with minimum
, all terms in the denominator approach
to zero, but the one with minimum
 approaches
to zero most slowly, and becomes the dominant term of the denominator.
If the numerator happens to be this term as well, then
approaches
to zero most slowly, and becomes the dominant term of the denominator.
If the numerator happens to be this term as well, then  ,
otherwise the numerator approaches zero and
,
otherwise the numerator approaches zero and  . Now
. Now  defined above becomes:
defined above becomes:
 |
(243) |
 defined in Eq.
(221) for a soft decision becomes a binary value
defined in Eq.
(221) for a soft decision becomes a binary value
 for a hard binary decision to assign
for a hard binary decision to assign
 to
to  with the smallest distance. Also
with the smallest distance. Also
 defined in Eq. (228)
as the sum of the posterior probabilities for all
defined in Eq. (228)
as the sum of the posterior probabilities for all  data
points to belong to
data
points to belong to  becomes
becomes  as the number of
data samples assigned only to
as the number of
data samples assigned only to  . In other words, now
the probabilistic EM method based on both
. In other words, now
the probabilistic EM method based on both  and
and
 becomes the deterministic K-means method based
on
becomes the deterministic K-means method based
on  only.
only.
We can also make a comparison between the GMM method for
unsupervised clustering and the softmax regression for
supervised classification. First, the latent variables
![${\bf Z}=[{\bf z}_1,\cdots,{\bf z}_N]$](img801.svg)
![${\bf Y}=[{\bf y}_1,\cdots,{\bf y}_N]$](img806.svg)




Examples
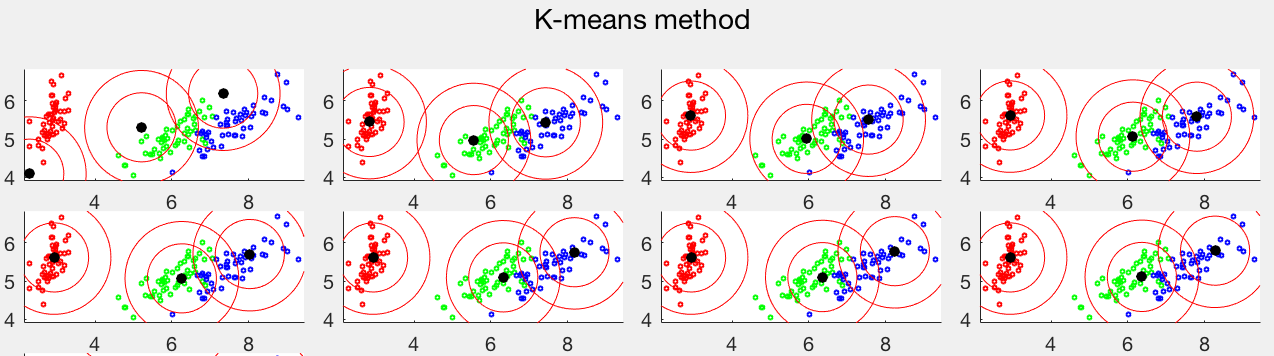
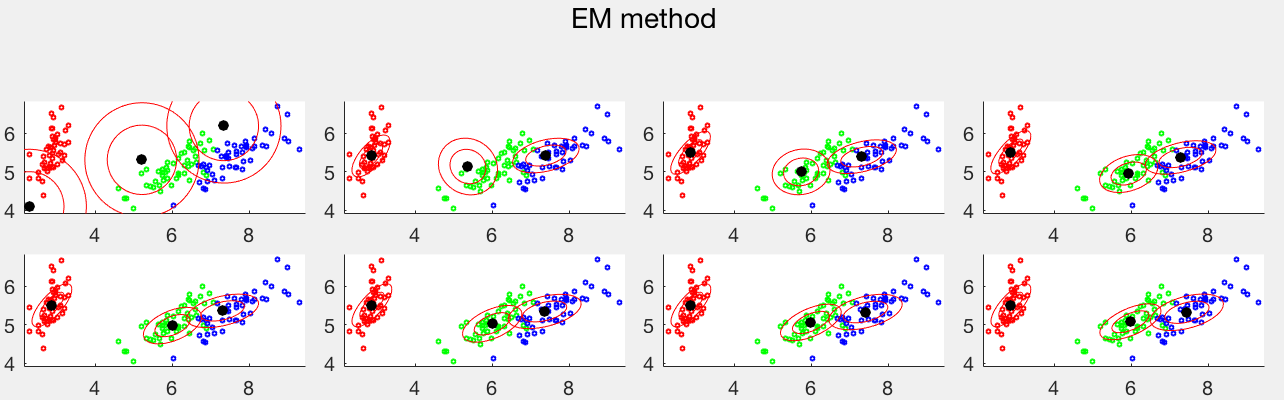
The same dataset is used to test both the K-means and EM clustering methods. The first panel shows 10 iterations of the K-means method, while the second panel shows 16 iterations of the EM method. In both cases, the iteration converges to the last plot. Comparing the two clustering results, we see that the K-means method cannot separate the red and green data points from two different clusters, both normally distributed with similar means but very different covariance matrices, while the blue data points all in the same cluster are separated into two clusters. But the EM method based on the Gaussian mixture model can correctly identified all three clusters.
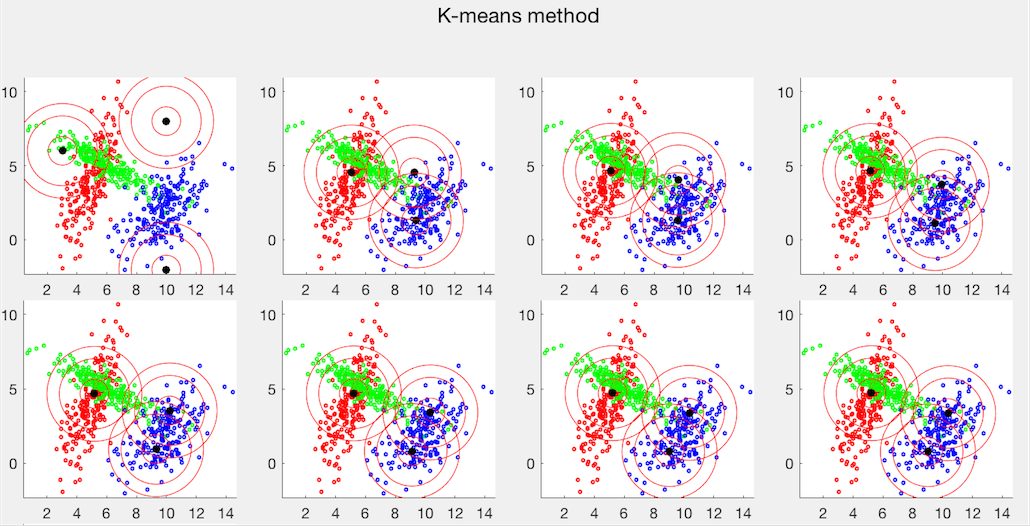
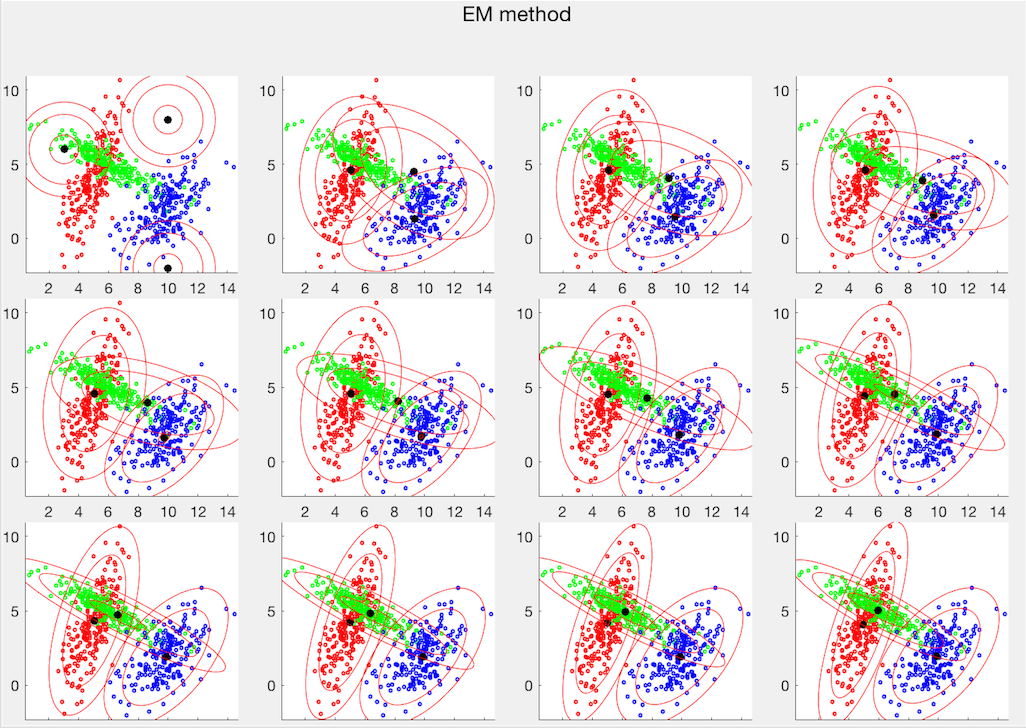
The two clustering methods are also applied to the Iris dataset, which has three classes each of 50 4-dimensional sample vectors. The PCA method is used to visualize the first two principal compnents, as shown below. Also, as can be seen from their c onfussion matrices, the error rate of the K-means method is 18/150, while that of the EM method is 5/150.
 |
(244) |