Next: Gaussian mixture model Up: Clustering Analysis Previous: Clustering Analysis
As suggested by the name of K-means clustering, in this method a set
of 


The K-means clustering algorithm can be formulated as an optimization problem to minimize an objective function
 |
(199) |
 |
(200) |
 is assigned to the kth cluster
is assigned to the kth cluster  if its
distance to the mean
if its
distance to the mean  is minimum. To minimize the objective
function
is minimum. To minimize the objective
function  with respective to
with respective to  , we set its derivative with
respect to each
, we set its derivative with
respect to each  to zero:
to zero:
 |
(201) |
 :
:
 |
(202) |
 is the number of all samples assigned to
cluster
is the number of all samples assigned to
cluster  , and the summation is over all samples assigned to
, and the summation is over all samples assigned to  .
.
As 




 clusters,
such as any
clusters,
such as any  samples of the dataset:
samples of the dataset:
 ,
set iteration index to zero
,
set iteration index to zero  ;
;
 in the dataset to one
of the
in the dataset to one
of the  clusters according to its distance to the corresponding
mean vector:
clusters according to its distance to the corresponding
mean vector:
if then then |
(203) |
 denotes the kth cluster with mean vector
denotes the kth cluster with mean vector
 in the lth iteration;
in the lth iteration;
 so that the objective function
so that the objective function  given above, i.e., the sum of
the distances squared from all
given above, i.e., the sum of
the distances squared from all
 to
to
 is minimized:
is minimized:
 |
(204) |
 |
(205) |
 , go back to Step 2.
, go back to Step 2.
This method is simple and effective, but it has the main drawback
that the number of clusters 



The Matlab code for the iteration loop of the algorithm is listed
below, where Mold and Mnew are respectively the 
Mnew=X(:,randi(N,1,K)); % use any K random samples as initial means
er=inf;
while er>0 % main iteration
Mold=Mnew;
Mnew=zeros(d,K); % initialize new means
Number=zeros(1,K);
for i=1:N % for all N samples
x=X(:,i);
dmin=inf;
for k=1:K % for all K clusters
d=norm(x-Mold(:,k));
if d<dmin
dmin=d; j=k;
end
end
Number(j)=Number(j)+1;
Mnew(:,j)=Mnew(:,j)+x;
end
for k=1:K
if Number(k)>0
Mnew(:,k)=Mnew(:,k)/Number(k);
end
end
er=norm(Mnew-Mold); % terminate if means no longer change
end
Example 1
The K-means algorithm is applied to a simulated dataset in 3-D space with





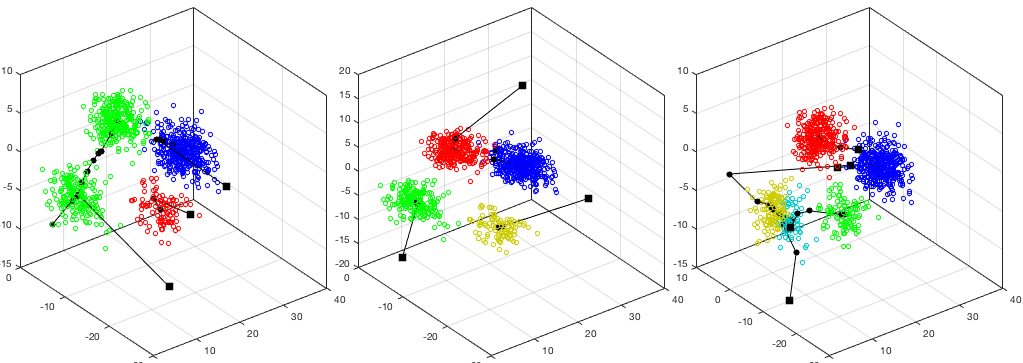
The clustering results corresponding to




 |
(206) |
 inter-cluster (Bhattacharyya) distances (between any
two of the
inter-cluster (Bhattacharyya) distances (between any
two of the  clusters):
clusters):
 |
(207) |
 , the intra-cluster distance of the 2nd cluster is
significantly greater than the other two, indicating the cluster may contain
two smaller clusters. Also, when
, the intra-cluster distance of the 2nd cluster is
significantly greater than the other two, indicating the cluster may contain
two smaller clusters. Also, when  , the inter-cluster distance between
clusters 3 and 4 is significantly smaller than others, indicating the two
clusters are too close and can therefore be merged.
, the inter-cluster distance between
clusters 3 and 4 is significantly smaller than others, indicating the two
clusters are too close and can therefore be merged.
While the K-means method is simple and effective, it has the main shortcoming
that the number of clusters 





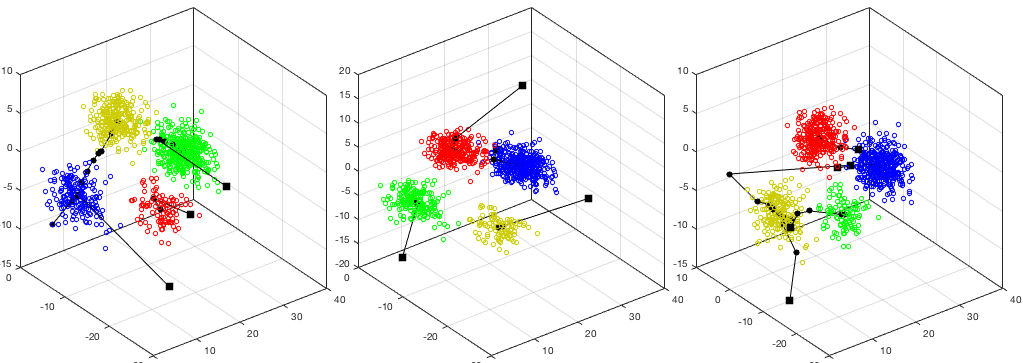
The idea of modifying the clustering results by merging and spliting leads to
the algorithm of Iterative Self-Organizing Data Analysis Technique (ISODATA),
which allows the number of clusters 
Example 2
The K-means clustering method is applied to the dataset of ten handwritten digits from 0 to 9 used previously. The clustering result is visualized based on the KLT that maps the data samples in the original 256-D space into the 3-D space spanned by the three eigenvectors corresponding to the three greatest eigenvalues of the covariance matrix of the dataset. The ground truth labelings are color coded as shown on the left, while the clustering result is shown on the right. We see that the clustering results match the original data reasonably well.
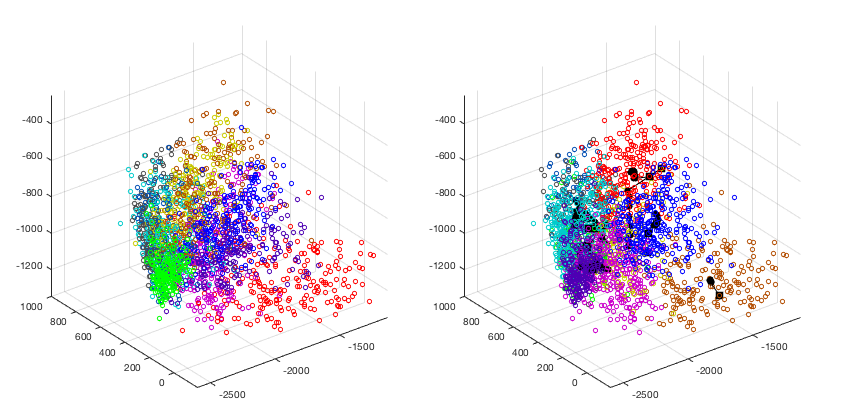
The clustering result is also shown in the confusion matrix, of which the columns and rows represent respectively the clusters based on the K-means clustering and the ground truth labeling (not used). In other words, the element in the ith row and jth column is the number of samples labeled to belong to the ith class but assigned to the jth cluster.
![$\displaystyle \left[ \begin{array}{rrrrrrrrrr}
1 & 0 & 25 & 165 & 27 & 2 & 3 & ...
...2 & 0 & 54 \\
4 & 2 & 0 & 1 & 0 & 28 & 119 & 1 & 64 & 5 \\
\end{array}\right]$](img777.svg) |
(208) |
All samples are converted from 256-dimensional vector back to