We revisit the linear regression problem but from the viewpoint
of Bayesian inference. Now the parameter  in the linear
regression model
in the linear
regression model
 is
assumed to be a random vector, and, as a function of
is
assumed to be a random vector, and, as a function of  ,
the regression function
,
the regression function
 is also random, and
so is the residual based on the training set
is also random, and
so is the residual based on the training set
 :
:
 i.e. i.e. |
(157) |
We further assume  has a zero-mean normal prior pdf:
has a zero-mean normal prior pdf:
 |
(158) |
based on the desire that the weights in  should take
values around zero, instead of large values on either positive
or negative, to avoid potiential overfitting. We also assume
the residual
should take
values around zero, instead of large values on either positive
or negative, to avoid potiential overfitting. We also assume
the residual  has a zero-mean normal pdf with a
diagonal covariance matrix, i.e., all components of
has a zero-mean normal pdf with a
diagonal covariance matrix, i.e., all components of  are indepenent of each other:
are indepenent of each other:
 |
(159) |
As a linear transformation of  , the regression function
, the regression function
 also has a zero-mean Gaussian
distribution, and so is
also has a zero-mean Gaussian
distribution, and so is
 as
the sum of two Gaussians.
as
the sum of two Gaussians.
To obtain the model parameter  , we consider its
likelihood
, we consider its
likelihood
 based on the
observed data
based on the
observed data
 :
:
This is the same Gaussian as
 ,
but shifted by
,
but shifted by
 .
Now we can find the optimal model parameter
.
Now we can find the optimal model parameter  by maximizing the likelihood
by maximizing the likelihood
 . This
method is called maximum likelihood estimation (MLE).
But as maximizing
. This
method is called maximum likelihood estimation (MLE).
But as maximizing
 is equivalent to
minimizing the SSE
is equivalent to
minimizing the SSE
 , the MLE method is
equivalent to the least squares method considered before.
, the MLE method is
equivalent to the least squares method considered before.
Maximizing the likelihood above is equivalent to minimizing
the negative log likelihood:
 |
(161) |
which is the same as the squared error
 defined in Eq. (114) to be minimized by
the LS method previously considered. We therefore see that the
least squares method and the maximum likelihood method are
equivalent to each other in this case.
defined in Eq. (114) to be minimized by
the LS method previously considered. We therefore see that the
least squares method and the maximum likelihood method are
equivalent to each other in this case.
We can further find the posterior
 of the
model parameter
of the
model parameter  given the observed data
given the observed data  ,
as the product of prior
,
as the product of prior
 and likelihood
and likelihood
 :
:
where all constant scaling factors (irrelavant to  ) are
dropped for simplicity. As both the prior and the likelihood are
Gaussian, this this posterior, when normalized, is also Gaussian
in terms of the mean
) are
dropped for simplicity. As both the prior and the likelihood are
Gaussian, this this posterior, when normalized, is also Gaussian
in terms of the mean
 and covariance
and covariance
 :
:
with the following covariance and mean:
Now we can find the optimal model parameter
 equal to the mean of the Gaussian
posterior where it is maximized. This solution is called the
maximum a posteriori (MAP) estimate of the model parameter
equal to the mean of the Gaussian
posterior where it is maximized. This solution is called the
maximum a posteriori (MAP) estimate of the model parameter
 .
.
We can now predict the function value of any unlabeled test
data point  based on the conditional probability
distribution
based on the conditional probability
distribution
 of the regression
model
of the regression
model
 , which,
as a linear transformation of the random parameters in
, which,
as a linear transformation of the random parameters in  ,
is also Gaussian
(see properties of Gaussians):
,
is also Gaussian
(see properties of Gaussians):
 |
(165) |
with the following mean and variance:
Furthermore, given a set of  unlabeled test data points in
unlabeled test data points in
![${\bf X}_*=[{\bf x}_{1*},\cdots,{\bf x}_{M*}]^T$](img815.svg) , we can find
, we can find
![$\displaystyle \hat{\bf y}_*={\bf f}_*={\bf f}({\bf x}_*)
=\left[\begin{array}{c...
...}\right]{\bf w}
=[{\bf x}_{1*},\cdots,{\bf x}_{M*}]^T{\bf w}={\bf X}^T_*{\bf w}$](img816.svg) |
(168) |
As a linear combination of Guassian  ,
,
 is also Gaussian:
is also Gaussian:
 |
(169) |
with the following mean and covariance:
The mean
![$\E[{\bf f}_*\vert{\bf X}_*,{\bf X},{\bf y} ]$](img823.svg) above
can be considered as the estimated function value
above
can be considered as the estimated function value
 at the test points in
at the test points in
 , while the variance
, while the variance
![$\Var[{\bf f}_*\vert{\bf X}_*,{\bf X},{\bf y}]$](img826.svg) as the confidence or certainty of the estimation.
as the confidence or certainty of the estimation.
Such a result produced by the Bayesian method in terms of the mean
and covariance of
 as a random vector is
quite different from the result produced by a frequentist method such
as the linear least square regression in terms of an estimated value
of
as a random vector is
quite different from the result produced by a frequentist method such
as the linear least square regression in terms of an estimated value
of
 as a deterministic variable. However, in the special
case of
as a deterministic variable. However, in the special
case of
 , i.e., the residual
, i.e., the residual  is no longer treated
as a random variable, then Eq. (170) above becomes the same
as Eq. (117) produced by linear least square regression, in
other words, the linear method method can be considered as a special
case of the Bayesian regression when the residual is deterministic.
is no longer treated
as a random variable, then Eq. (170) above becomes the same
as Eq. (117) produced by linear least square regression, in
other words, the linear method method can be considered as a special
case of the Bayesian regression when the residual is deterministic.
The method considered above is based on the linear regression model
 , but it can also be generalized to
nonlinear functions, same as in the case of
generalized linear regression
discussed previously. To do so, the regression function is assumed
to be a linear combination of a set of
, but it can also be generalized to
nonlinear functions, same as in the case of
generalized linear regression
discussed previously. To do so, the regression function is assumed
to be a linear combination of a set of  nonlinear mapping functions
nonlinear mapping functions
 :
:
 |
(172) |
where
![$\displaystyle {\bf w}=[w_1,\cdots,w_K]^T,\;\;\;\;\;\;
{\bf\phi}({\bf x})=[\varphi_1({\bf x}),\cdots,\varphi_K({\bf x})]^T$](img833.svg) |
(173) |
For example, some commenly used mapping functions are listed in Eq.
(155).
Applying this regression model to each of the  observed data
points in
observed data
points in
 , we
get
, we
get
 with residual
with residual
 , which can be written in matrix form:
, which can be written in matrix form:
where, as previously defined in Eq. (152),
![$\displaystyle {\bf\phi}_n={\bf\phi}({\bf x}_n)=\left[\begin{array}{c}
\varphi_1...
..._n)
\end{array}\right],
\;\;\;\;\;\;
{\bf\Phi}=[{\bf\phi}_1,\cdots,{\bf\phi}_N]$](img840.svg) |
(175) |
As this nonlinear model
 is in the
same form as the linear model
is in the
same form as the linear model
 , all
previous discussion for the linear Bayesian regression is still
valid for this nonlinear model, if
, all
previous discussion for the linear Bayesian regression is still
valid for this nonlinear model, if
![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]_{(d+1)\times N}$](img841.svg) is replaced
by
is replaced
by
![${\bf\Phi}=[{\bf\phi}_1,\cdots,{\bf\phi}_N]_{K\times N}$](img842.svg) .
We can get the covariance matrix and mean vector of the posterior
.
We can get the covariance matrix and mean vector of the posterior
 in the K-dimensional space based on the
observed data
in the K-dimensional space based on the
observed data
 :
:
The function
 of the multiple
test points in
of the multiple
test points in
![${\bf X}_*=[{\bf x}_{*1},\cdots,{\bf x}_{*M}]^T$](img846.svg) is
a linear transformation of
is
a linear transformation of  :
:
![$\displaystyle \hat{\bf y}_*={\bf f}_*={\bf f}({\bf x}_*)
=\left[\begin{array}{c...
...ight]_*{\bf w}
=[{\bf\phi}_1,\cdots,{\bf\phi}_M]^T{\bf w}
={\bf\Phi}^T_*{\bf w}$](img847.svg) |
(177) |
and it also has a Gaussian joint distribution:
 |
(178) |
with the mean and covariance:
They can be reformulated to become:
and
where we have used Theorem 1 in the Appendices.
We note that in these expressions the mapping function
 only shows up in the quadratic form of
only shows up in the quadratic form of
 ,
,
 ,
or
,
or
 . This suggests that we
may consider using
kernel method, by
which some significant advantage may be gained (such as in the
support-vector machines).
Specifically, if we replace the mn-th component
. This suggests that we
may consider using
kernel method, by
which some significant advantage may be gained (such as in the
support-vector machines).
Specifically, if we replace the mn-th component
 of
of
 by a kernel function
only in terms of
by a kernel function
only in terms of  and
and  :
:
 |
(182) |
then the mapping function
 no longer needs to
be explicitly specified. This idea is called the kernel trick,
Now we have
no longer needs to
be explicitly specified. This idea is called the kernel trick,
Now we have
![$\displaystyle {\bf\Phi}^T{\bf\Sigma}_w{\bf\Phi}
=\left[\begin{array}{c}{\bf\phi...
...k({\bf x}_N,{\bf x}_1)&\cdots&k({\bf x}_N,{\bf x}_N)\end{array}\right]
={\bf K}$](img865.svg) |
(183) |
and similarly we also have
 and
and
 . These kernel
functions are symmetric, same as all covariance matrices. Now the mean
and covariance of
. These kernel
functions are symmetric, same as all covariance matrices. Now the mean
and covariance of  given above can be expressed as:
given above can be expressed as:
As a general summary of the method discussed above, we note that it is
essentially different from the frequentist approach, in the sense that
the estimated result produced is no longer a set specific values of some
unknown variables in question (function values, parameters of regression
models, etc.), but the probability distribution of these variables in terms
of the expectation and covariance. Consequently, we can not only treat the
expectation as the estimated values, but also use the variance as the
confidence or certainty of such an estimation.
Below is a segment of Matlab code for estimating the mean and covariance
of weight vector  by the Bayes linear regression. Here
by the Bayes linear regression. Here X
and y are for  and
and  of the training data
of the training data  ,
and
,
and Xstar, Phi and Phistar are for  ,
,
 and
and
 , respectively.
, respectively.
SigmaP=eye(d); % prior variance of r in original space
SigmaQ=eye(D); % prior variance of r in feature space
Sigmaw1=inv(X*X'/sigmaN^2+inv(SigmaP)); % covariance of w by Bayes regression
Sigmaw2=inv(Phi0*Phi0'/sigmaN^2+inv(SigmaQ)); % covariance of w with basis function
w0=inv(X*X')*X*y % weight vector w by LS method
w1=Sigmaw1*X*y/sigmaN^2 % mean of w by Bayes regression
w2=Sigmaw2*Phi0*y/sigmaN^2; % mean of w vector by Bayes regression with basis function
y0=w0'*Xstar; % y by least squares method
y1=w1'*Xstar; % y by Bayes regression
y2=w2'*Phistar0; % y by Bayes regression with basis function
for i=1:M
sgm1(i)=sqrt(Xstar(:,i)'*Sigmaw1*Xstar(:,i)); % variance of estimated y
sgm2(i)=sqrt(Phistar(:,i)'*Sigmaw2*Phistar(:,i));
end
Example
Consider a 2-D linear function
![$\displaystyle f({\bf x})={\bf w}^T{\bf x}=[w_0,\,w_1]\left[\begin{array}{c}x_0\\ x_1\end{array}\right]
=w_0x_0+w_1x_1=0.5+0.5\,x_1$](img874.svg) |
(185) |
where we have assumed  ,
,  and
and  , i.e., the
function is actually a straight line
, i.e., the
function is actually a straight line
 with intercept
with intercept
 and slope
and slope  . The distribution of the additive noise
(residual) is
. The distribution of the additive noise
(residual) is
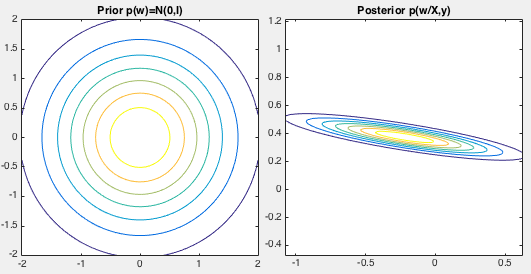
 . The prior distributions
of the weights
. The prior distributions
of the weights
![${\bf w}=[w_0,w_1]^T$](img879.svg) is assumed to be
is assumed to be
 , and posterior
, and posterior
 can be obtained based on the observed data
can be obtained based on the observed data
 ,
both shown in the figure below. We see that the posterior is concentrated
around
,
both shown in the figure below. We see that the posterior is concentrated
around
 and
and
 , the estimates of the true
weights
, the estimates of the true
weights  and
and  , based on the observed noisy data.
, based on the observed noisy data.
Various regression methods including both the least-squared method and the
Bayesian linear regression methods, and in feature space spanned by a set
of basis functions
 based on each of
the three functions given above are carried out with their results shown
in the figure below:
based on each of
the three functions given above are carried out with their results shown
in the figure below:
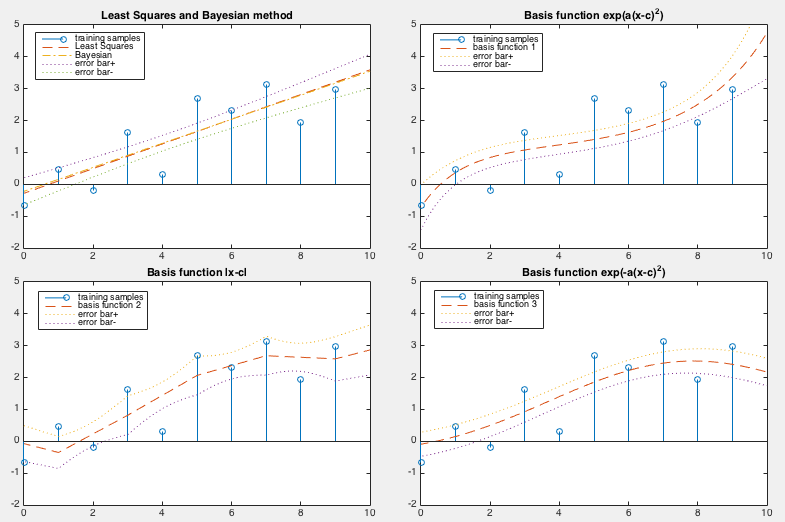
The LS errors for these methods are listed below:
 |
(186) |
We see that the linear methods (the pseudo inverse and the Bayesian
regression) produce similar estimated weights error, but smaller error
can be achieved by the same method of Bayesian linear regression if
proper nonlinear mapping is used (e.g., the last two mapping functions).





 i.e.
i.e.
 has a zero-mean normal prior pdf:
has a zero-mean normal prior pdf:

 should take
values around zero, instead of large values on either positive
or negative, to avoid potiential overfitting. We also assume
the residual
should take
values around zero, instead of large values on either positive
or negative, to avoid potiential overfitting. We also assume
the residual  has a zero-mean normal pdf with a
diagonal covariance matrix, i.e., all components of
has a zero-mean normal pdf with a
diagonal covariance matrix, i.e., all components of  are indepenent of each other:
are indepenent of each other:

 , the regression function
, the regression function
 also has a zero-mean Gaussian
distribution, and so is
also has a zero-mean Gaussian
distribution, and so is
 as
the sum of two Gaussians.
as
the sum of two Gaussians.



 ,
but shifted by
,
but shifted by
 .
Now we can find the optimal model parameter
.
Now we can find the optimal model parameter  by maximizing the likelihood
by maximizing the likelihood
 . This
method is called maximum likelihood estimation (MLE).
But as maximizing
. This
method is called maximum likelihood estimation (MLE).
But as maximizing
 is equivalent to
minimizing the SSE
is equivalent to
minimizing the SSE
 , the MLE method is
equivalent to the least squares method considered before.
, the MLE method is
equivalent to the least squares method considered before.

 defined in Eq. (114) to be minimized by
the LS method previously considered. We therefore see that the
least squares method and the maximum likelihood method are
equivalent to each other in this case.
defined in Eq. (114) to be minimized by
the LS method previously considered. We therefore see that the
least squares method and the maximum likelihood method are
equivalent to each other in this case.





 ) are
dropped for simplicity. As both the prior and the likelihood are
Gaussian, this this posterior, when normalized, is also Gaussian
in terms of the mean
) are
dropped for simplicity. As both the prior and the likelihood are
Gaussian, this this posterior, when normalized, is also Gaussian
in terms of the mean
 and covariance
and covariance
 :
:


![$\displaystyle \exp \left[ -\frac{1}{2} \left(
({\bf w}-{\bf m}_{w/d})^T {\bf\Sigma}_{w/d}^{-1}({\bf w}-{\bf m}_{w/d})
\right) \right]$](img800.svg)

![$\displaystyle \exp \left[ -\frac{1}{2} \left(
{\bf w}^T{\bf\Sigma}_{w/d}^{-1}{\bf w}-2{\bf m}_{w/d}^T {\bf\Sigma}_{w/d}^{-1}{\bf w}
\right) \right]$](img801.svg)






 equal to the mean of the Gaussian
posterior where it is maximized. This solution is called the
maximum a posteriori (MAP) estimate of the model parameter
equal to the mean of the Gaussian
posterior where it is maximized. This solution is called the
maximum a posteriori (MAP) estimate of the model parameter
 .
.





![$\displaystyle \E[f_*\vert{\bf x}_*,{\bf X},{\bf y}]$](img811.svg)


![$\displaystyle \Var[f_*\vert{\bf x}_*,{\bf X},{\bf y}]$](img813.svg)



![${\bf X}_*=[{\bf x}_{1*},\cdots,{\bf x}_{M*}]^T$](img815.svg)
![$\displaystyle \hat{\bf y}_*={\bf f}_*={\bf f}({\bf x}_*)
=\left[\begin{array}{c...
...}\right]{\bf w}
=[{\bf x}_{1*},\cdots,{\bf x}_{M*}]^T{\bf w}={\bf X}^T_*{\bf w}$](img816.svg)
 ,
,
 is also Gaussian:
is also Gaussian:

![$\E[{\bf f}_*\vert{\bf X}_*,{\bf X},{\bf y} ]$](img823.svg) above
can be considered as the estimated function value
above
can be considered as the estimated function value
 at the test points in
at the test points in
 , while the variance
, while the variance
![$\Var[{\bf f}_*\vert{\bf X}_*,{\bf X},{\bf y}]$](img826.svg) as the confidence or certainty of the estimation.
as the confidence or certainty of the estimation.








![$\displaystyle {\bf w}=[w_1,\cdots,w_K]^T,\;\;\;\;\;\;
{\bf\phi}({\bf x})=[\varphi_1({\bf x}),\cdots,\varphi_K({\bf x})]^T$](img833.svg)






![$\displaystyle \left[\begin{array}{c}y_1\\ \vdots\\ y_N\end{array}\right]_{N\tim...
...imes 1}
+\left[\begin{array}{c}r_1\\ \vdots\\ r_N\end{array}\right]_{N\times 1}$](img838.svg)

![$\displaystyle \left[\begin{array}{c}
{\bf\phi}^T_1\\ \vdots\\ {\bf\phi}^T_N
\end{array}\right]{\bf w}+{\bf r}={\bf\Phi}^T{\bf w}+{\bf r}$](img839.svg)
![$\displaystyle {\bf\phi}_n={\bf\phi}({\bf x}_n)=\left[\begin{array}{c}
\varphi_1...
..._n)
\end{array}\right],
\;\;\;\;\;\;
{\bf\Phi}=[{\bf\phi}_1,\cdots,{\bf\phi}_N]$](img840.svg)


![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]_{(d+1)\times N}$](img841.svg)
![${\bf\Phi}=[{\bf\phi}_1,\cdots,{\bf\phi}_N]_{K\times N}$](img842.svg)








 of the multiple
test points in
of the multiple
test points in
![${\bf X}_*=[{\bf x}_{*1},\cdots,{\bf x}_{*M}]^T$](img846.svg) is
a linear transformation of
is
a linear transformation of  :
:
![$\displaystyle \hat{\bf y}_*={\bf f}_*={\bf f}({\bf x}_*)
=\left[\begin{array}{c...
...ight]_*{\bf w}
=[{\bf\phi}_1,\cdots,{\bf\phi}_M]^T{\bf w}
={\bf\Phi}^T_*{\bf w}$](img847.svg)

![$\displaystyle \E[ {\bf f}_*\vert{\bf X}_*,{\bf X},{\bf y} ]$](img819.svg)

![$\displaystyle \E[ {\bf\Phi}^T_*{\bf w} ]={\bf\Phi}^T_*\E[ {\bf w} ]
={\bf\Phi}^T_*{\bf m}_{w/d}$](img849.svg)


![$\displaystyle \E[ {\bf f}_*{\bf f}_*^T ]
=\E[ {\bf\Phi}^T_*{\bf w} {\bf w}^T{\b...
...*\E[ {\bf w} {\bf w}^T ] {\bf\Phi}_*
={\bf\Phi}^T_*{\bf\Sigma}_{w/d}{\bf\Phi}_*$](img850.svg)
![$\displaystyle \E[ {\bf f}_*\vert{\bf X}_*,{\bf X},{\bf y} ]$](img819.svg)





![$\displaystyle {\bf\Phi}^T_*{\bf\Sigma}_w{\bf\Phi}
\left[{\bf\Phi}^{-1}\left({\b...
...+\sigma_r^2{\bf\Sigma}_w^{-1}\right){\bf\Sigma}_w{\bf\Phi} \right]^{-1} {\bf y}$](img853.svg)






![$\displaystyle {\bf\Phi}^T_*\left[{\bf\Sigma}_w-{\bf\Sigma}_w{\bf\Phi}
({\bf\Phi...
...a}_w{\bf\Phi}+\sigma_r^2{\bf I})^{-1}{\bf\Phi}^T{\bf\Sigma}_w\right]{\bf\Phi}_*$](img856.svg)











 no longer needs to
be explicitly specified. This idea is called the kernel trick,
Now we have
no longer needs to
be explicitly specified. This idea is called the kernel trick,
Now we have
![$\displaystyle {\bf\Phi}^T{\bf\Sigma}_w{\bf\Phi}
=\left[\begin{array}{c}{\bf\phi...
...k({\bf x}_N,{\bf x}_1)&\cdots&k({\bf x}_N,{\bf x}_N)\end{array}\right]
={\bf K}$](img865.svg)
 and
and
 . These kernel
functions are symmetric, same as all covariance matrices. Now the mean
and covariance of
. These kernel
functions are symmetric, same as all covariance matrices. Now the mean
and covariance of  given above can be expressed as:
given above can be expressed as:







![$\displaystyle f({\bf x})={\bf w}^T{\bf x}=[w_0,\,w_1]\left[\begin{array}{c}x_0\\ x_1\end{array}\right]
=w_0x_0+w_1x_1=0.5+0.5\,x_1$](img874.svg)
 ,
,  and
and  , i.e., the
function is actually a straight line
, i.e., the
function is actually a straight line
 with intercept
with intercept
 and slope
and slope  . The distribution of the additive noise
(residual) is
. The distribution of the additive noise
(residual) is
 . The prior distributions
of the weights
. The prior distributions
of the weights
![${\bf w}=[w_0,w_1]^T$](img879.svg) is assumed to be
is assumed to be
 , and posterior
, and posterior
 can be obtained based on the observed data
can be obtained based on the observed data
 ,
both shown in the figure below. We see that the posterior is concentrated
around
,
both shown in the figure below. We see that the posterior is concentrated
around
 and
and
 , the estimates of the true
weights
, the estimates of the true
weights  and
and  , based on the observed noisy data.
, based on the observed noisy data.



