Next: About this document ... Up: Regression Analysis and Classification Previous: Gaussian Process Classifier -
The binary GPC considered previously can be generalized to multi-class
GPC based on softmax function, similar to the how binary classification
based on logistic function is generalized to multi-class classification.
First of all, we define the following variables for each class



Binary labeling
![${\bf y}_k=[y_{1k},\cdots,y_{Nk}]^T$](img1366.svg)

![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img498.svg)

 |
(297) |
 is
labeled by
is
labeled by  binary variables
binary variables  and
and  for
all
for
all
 (same as in softmax regression),
as well as an integer labeling
(same as in softmax regression),
as well as an integer labeling  .
.
Latent function
![${\bf f}_k=[f_{1k},\cdots,f_{Nk}]^T$](img1369.svg)



![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img498.svg)
Probability
![${\bf p}_k=[p_{1k},\cdots,p_{Nk}]^T$](img1371.svg)



 then necessarily
then necessarily  for all
for all
 but
but  , i.e., the
, i.e., the  variables
variables
 are not independent.
given
are not independent.
given
 , we do not need to consider
, we do not need to consider
 for any
for any  .
.
The probability for 



 |
(299) |
Based on 




![$\displaystyle {\bf y}=\left[\begin{array}{c}{\bf y}_1\\ \vdots\\ {\bf y}_K\end{...
...p_{11}\\ \vdots\\ p_{N1}\\ \vdots\\ p_{1K}\\ \vdots\\ p_{NK}
\end{array}\right]$](img1385.svg) |
(300) |
The posterior of 

 |
(301) |
We first find the likelihood based on all 
 |
(302) |
 belongs to only one of the
belongs to only one of the  classes,
only one of
classes,
only one of
 can be 1 while all others are
0, consequently, the product above contains only
can be 1 while all others are
0, consequently, the product above contains only  probabilities
raised to the power of
probabilities
raised to the power of  , each for one of the
, each for one of the  samples
belonging to a certain class, while all other probabilities raised
to the power of
samples
belonging to a certain class, while all other probabilities raised
to the power of  become 1 and not considered.
become 1 and not considered.
We next assume the prior probability of the latent function




 |
(303) |
 containing all
containing all  such latent functions
is also a Gaussian
such latent functions
is also a Gaussian
 ,
where
,
where  is a KN-dimensional zero vector and
is a KN-dimensional zero vector and  is a
is a
 block diagonal matrix
block diagonal matrix
![$\displaystyle {\bf K}=\left[\begin{array}{cccc}{\bf K}_1 & {\bf0} &\cdots &{\bf...
...ots & \ddots & \vdots \\
{\bf0} & \cdots & {\bf0} &{\bf K}_K\end{array}\right]$](img1397.svg) |
(304) |
 as defined in
Eq. (254) on the diagonal. All off-diagonal
blocks are zero as the latent functions of different classes are
uncorrelated.
as defined in
Eq. (254) on the diagonal. All off-diagonal
blocks are zero as the latent functions of different classes are
uncorrelated.
Having found both the likelihood


 |
(305) |
 is not Gaussian,
as
is not Gaussian,
as
 is a binary labeling, instead of a
continuous variable. As a product of the Gaussian prior and
non-Gaussian likelihood, the posterior
is a binary labeling, instead of a
continuous variable. As a product of the Gaussian prior and
non-Gaussian likelihood, the posterior
 is
not a Gaussian process either. However, for convnience, we still
assume it is approximately Gaussian
is
not a Gaussian process either. However, for convnience, we still
assume it is approximately Gaussian
 , of which
the mean
, of which
the mean
 and covariance
and covariance
 are to
be found.
are to
be found.
Taking log of the posterior, we get
 |
 |
 |
|
 |
 |
(306) |
 . The
gradient of
. The
gradient of
 is
is
 |
 |
 |
|
 |
 |
(307) |
 comes from
Note that
comes from
Note that
 is a function of
is a function of  .
.
As the posterior





 |
(309) |
 is the vector defined
above evaluated at
is the vector defined
above evaluated at
 .
.
We further get the Hessian matrix of

 |
(311) |
 :
Here
:
Here
 is a
is a
 Jacobian matrix
of
Jacobian matrix
of  , of which the
, of which the  -th component
-th component
 is:
is:
 |
 |
 |
|
 |
![$\displaystyle \left[\frac{e^{f_{ik}}}{\sum_{h=1}^K e^{f_{ih}}}\delta_{kl}
-\fra...
...h}}\right)^2}\right]
\delta_{ij}
=(p_{ik}\delta_{kl}-p_{ik} p_{jl})\delta_{ij},$](img1422.svg) |
(313) |
 can be written also in two terms:
where the first term
can be written also in two terms:
where the first term
 is a diagnal matrix containing
all
is a diagnal matrix containing
all  components of
components of  along the diagnal (
along the diagnal ( and
and  ),
and
),
and  in the second term is a KN by N matrix composed of
in the second term is a KN by N matrix composed of 
 diagonal matrices
diagonal matrices
 :
:
![$\displaystyle {\bf P}=\left[\begin{array}{c}diag({\bf p}_1)\\
\vdots\\ diag({\...
...& \vdots\\
0 & \cdots & 0 & p_{Nk}
\end{array}\right],\;\;\;\;\;(k=1,\cdots,K)$](img1428.svg) |
(315) |
![$\displaystyle {\bf PP}^T=\left[\begin{array}{c}diag({\bf p}_1)\\
\vdots\\ diag...
...diag({\bf p}_K) diag({\bf p}_1) & \cdots & diag({\bf p}_K^2)
\end{array}\right]$](img1429.svg) |
(316) |
![$\displaystyle diag({\bf p}_k) diag({\bf p}_l)=\left[\begin{array}{cccc}
p_1^kp_...
...& \vdots & \ddots & \vdots \\
0 & \cdots & 0 & p_{Nk}p_{Nl} \end{array}\right]$](img1430.svg) |
(317) |
Having found both the gradient




 |
 |
 |
|
 |
 |
||
 |
 |
(318) |
 , but also
, but also  as a functions of
as a functions of  given
in Eq. (308).
given
in Eq. (308).
Now that we have got both






 :
:
Similar to Eq. (292) in the binary case, we have
the following based on

 |
 |
 |
|
 |
![$\displaystyle {\bf K}_*^T{\bf K}^{-1}\,[{\bf K}({\bf y}-{\bf p}_{m_f})]
={\bf K}_*^T({\bf y}-{\bf p}_{m_f})$](img1438.svg) |
(319) |
 classes are uncorrelated, i.e.,
classes are uncorrelated, i.e.,  is block-diagonal,
the above can be separated into
is block-diagonal,
the above can be separated into  equations each for one of the classes:
equations each for one of the classes:
 |
(320) |
 :
:
Similar to Eq. (294) in the binary case, we have
the following based on

 |
(321) |
 is given in Eq. (314)
with
is given in Eq. (314)
with  evaluated at
evaluated at
 . Then, similar to
Eq. (294), we get
. Then, similar to
Eq. (294), we get
 |
(322) |
Now we can further get the probability for 

 |
(323) |
 to class
to class  if
if
 .
.
Moreover, the certainty or confidence of this classification result
can be found from

The Matlab code for the essential parts of the algorithm is
listed below. First, the following code segment carries out the
classification of 


K=Kernel(X,X); % covariance of prior of p(f|X)
Ks=Kernel(X,Xs);
Kss=Kernel(Xs,Xs);
[meanf Sigmaf p W]=findPosteriorMeanMC(K,y,C);
% find mean and covariance of p(f|D), and W, p
Sigmafy=Kss-Ks'*inv(K+inv(W))*Ks; % covariance of p(f|D)
p=reshape(p,N,C);
y=reshape(y,N,C);
for k=1:C
meanfD(:,k)=Ks'*(y(:,k)-p(:,k); % mean of p(f_*|X_*,D) for kth class
end
for i=1:n % for each of n test samples
d=sum(exp(meanfD(i,:))); % denominator of softmax function
[pmax k]=max(exp(meanfD(i,:))/d); % find class with max probability
ys(i)=k; % label ith sample as member of kth class
pr(i,k)=pmax; % probility of ith sample belonging to kth class
end
The code segment above calls the following function which computes the
mean and covariance of

function [meanf Sigmaf p W]=findPosteriorMeanMC(K,y,C)
% find mean and covariance of p(f|X,y) by Newton's method, based on D={X,y}
n=length(y); % number of training samples
f0=zeros(n,1); % initial value of latent function
f=zeros(n,1);
er=1;
k=0;
while er > 10^(-9) % find f that maximizes p(f|X,y)
k=k+1;
[p W]=findp(f,N,C); % call function findp to find vector p and matrix W
f=inv(inv(K)+W)*(W*f0+y-p); % iteratively update value of f
er=norm(f-f0); % difference between consecutive iterations
f0=f; % update f
end
meanf=f;
Sigmaf=inv(inv(K)+W);
end
The following function called by the previous function finds vector


function [p W]=findp(f,N,C) % find vector p and matrix W=diag(p)-P*P'
F=reshape(f,N,C)'; % kth row contains N samples of class k
p=zeros(C,N); % initialize p
for n=1:N % for each of N training samples
d=sum(exp(F(:,n))); % sum of all C terms in denominator
for k=1:C % for all C classes
p(k,n)=exp(F(k,n))/d;
end
end
P=[];
for k=1:C % generate P
P=[P; diag(p(k,:))]; % stack C diagonal matrices
end
p=reshape(p',N*C,1); % convert p into a column vector
W=diag(p)-P*P'; % generate W matrix
end
Example 1:
This example shows the classification result of the same dataset of
three classes used before. The top two panels show the distributions
of the three classes in the training data set. The bottom two panels
show the classification results in terms of the partitioning of the
feature space (bottom left) and the posterior distribution


![$\displaystyle \left[\begin{array}{rrr}
197 & 3 & 0 \\
0 & 195 & 5 \\
1 & 21 & 178 \\
\end{array}\right]$](img1448.svg) |
(324) |
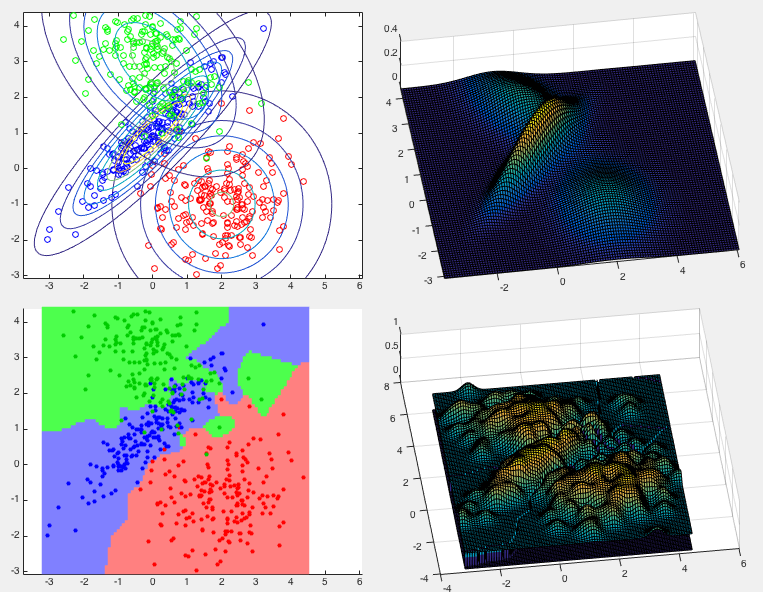
Example 2:
This example shows the classification of the XOR data set. The confusion
matrix of the classification result is shown below, with the error rate

![$\displaystyle \left[\begin{array}{rr}
193 & 7 \\
6 & 194
\end{array}\right]$](img1450.svg) |
(325) |
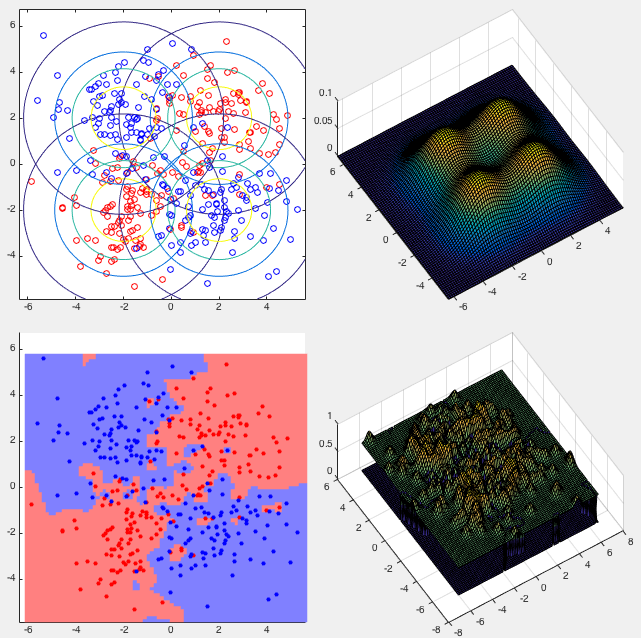
We see that in both examples, the error rates of the GPC method are lower than those of the naive Bayesian method. However, the naive Bayesian method does not have the overfitting problem, while in the method of GPC, we may need to carefully adjust the parameter of squared exponential for the kernel functions to make proper tradeoff between overfitting and error rate.