Next: Gaussian Process Classifier - Up: Regression Analysis and Classification Previous: Gaussian Process Regression
In both logistic regression and softmax regression considered
previously, we convert the linear regression function






Here in binary GPC, we assume the training samples are labeled
by 1 or -1 (instead of 0) for mathematical convenience, i.e.,











![$\displaystyle p(y=1\vert{\bf x},{\cal D}) = E_f[\sigma(f({\bf x}))]
=\int\sigma(f({\bf x}))\;p(f\vert{\bf x},{\cal D})\,d f$](img1251.svg) |
(265) |
 is marginalized (averaged out) in the
integral above, it is hidden instead of explicitly specified,
and is therefore called a latent function.
is marginalized (averaged out) in the
integral above, it is hidden instead of explicitly specified,
and is therefore called a latent function.
For the training set
![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img498.svg)
![${\bf f}({\bf X})=[f({\bf x}_1),\cdots,f({\bf x})]^T$](img1252.svg)
 is the posterior which can be found
in terms of the likelihood
is the posterior which can be found
in terms of the likelihood
 and the prior
and the prior
 based on
Bayes' theorem:
As always, the denominator
based on
Bayes' theorem:
As always, the denominator
 independent of
independent of
 is dropped.
is dropped.
We first find the likelihood






 |
 |
 |
|
 |
 |
 |
(268) |
 given
given
 :
The likelihood of
:
The likelihood of  for all
for all  i.i.d. samples in the
training set
i.i.d. samples in the
training set  is:
is:
 |
(270) |
We then consider the prior







 |
(271) |
 and
and
 are more correlated if
are more correlated if
 and
and  are close together, but less so if
they are farther apart. However the justification for such a
property is different. In GPR, this property is desired for the
smoothness of the regression function; while here in GPC, this
property is also desired so that function values
are close together, but less so if
they are farther apart. However the justification for such a
property is different. In GPR, this property is desired for the
smoothness of the regression function; while here in GPC, this
property is also desired so that function values
 and
and
 for two samples
for two samples  and
and  close to each other in the feature space are more correlated and
therefore the two samples are more likely to be classified into
the same class, but less so if they are far apart.
close to each other in the feature space are more correlated and
therefore the two samples are more likely to be classified into
the same class, but less so if they are far apart.
Also, as discussed in GPR, here the parameter 









Having found both the likelihood


 is not Gaussian, as
the binary labeling
is not Gaussian, as
the binary labeling  of the training set
of the training set  is not
continuous. Consequently, the posterior
is not
continuous. Consequently, the posterior
 , as a
product of the Gaussian prior and non-Gaussian likelihood, is not
Gaussian. However, we can carry out Laplace approximation
and still approximate the posterior as a Gaussian:
, as a
product of the Gaussian prior and non-Gaussian likelihood, is not
Gaussian. However, we can carry out Laplace approximation
and still approximate the posterior as a Gaussian:
 |
(273) |
 and covariance
and covariance
 are to be obtained in the following. We further get the log posterior
denoted by
are to be obtained in the following. We further get the log posterior
denoted by
 :
The two middle terms are constant independent of
:
The two middle terms are constant independent of  and can therefore be dropped.
and can therefore be dropped.
Now that the posterior



On the other hand, we can also find


 |
(280) |
 of
of  :
:
 |
 |
 |
|
 |
 |
 |
(281) |
 and
and  . We then
find
and
. We then
find
and
Equatiing the two expressions for

 in
Eqs. (275) and (277) we get:
in
Eqs. (275) and (277) we get:
 i.e., i.e., |
(285) |
 given in Eq. (284) into
the equation and solving it for
given in Eq. (284) into
the equation and solving it for
 , we get
, we get
 |
(286) |
 , in consistence
with the fact that when
, in consistence
with the fact that when
 , the log posterior
, the log posterior
 , assumed to be Gaussian,
reaches maximum with
, assumed to be Gaussian,
reaches maximum with
 . However,
we note that in the equation above,
. However,
we note that in the equation above,
 as a value of
as a value of
 is expressed in terms of both
is expressed in terms of both  and
and  ,
which in turn are functions of
,
which in turn are functions of  as given in
Eqs. (282) and (283), i.e.,
as given in
Eqs. (282) and (283), i.e.,
 given
above is not in a closed form, as
given
above is not in a closed form, as  and parameters
and parameters
 and
and  are interdependent. We instead need to
carry out an iteration during which both
are interdependent. We instead need to
carry out an iteration during which both  and the
parameters
and the
parameters  and
and  are updated alternatively:
are updated alternatively:
 ,
at which
,
at which
 is maximized, and
Now that
is maximized, and
Now that  and
and  as well as
as well as
 are available, we can further obtain
are available, we can further obtain
 in
Eq. (284), and the posterior
in
Eq. (284), and the posterior
 .
.
Having found


 |
(289) |
 in the equation as a Gaussian:
in the equation as a Gaussian:
 |
(290) |
 and covariance
and covariance
 can
be obtained based on the fact that both
can
be obtained based on the fact that both
 and
and
 are the same Gaussian process, i.e.,
their joint probability
are the same Gaussian process, i.e.,
their joint probability
 is a Gaussian. The
method is therefore the same as what is discussed in the method of
GPR. Specifically, we take
the following steps:
is a Gaussian. The
method is therefore the same as what is discussed in the method of
GPR. Specifically, we take
the following steps:
 and
covariance
and
covariance
 of
of
 conditioned
on the latent function
conditioned
on the latent function  , same as in Eq. (258)
for GPR:
, same as in Eq. (258)
for GPR:
 as the expectation of
as the expectation of
 conditioned on
conditioned on
 , i.e. find the average of
, i.e. find the average of
 over
over  based on
based on
 (marginalize over
(marginalize over  ):
):
 given in
Eq. (288).
given in
Eq. (288).
![${\bf\Sigma}_{m_{f_*\vert f}}=E_f[({\bf m}_{f_*\vert f}-{\bf m}_{f_*})^2]$](img1333.svg) .
Given the covariance
.
Given the covariance
 of
of  in Eq. (284), we can further find the covariance of
in Eq. (284), we can further find the covariance of
 as a linear combination
of
as a linear combination
of  (Recall if
(Recall if
 , then
, then
 ):
):
 |
(293) |
 of
of  as the sum of
as the sum of
![${\bf\Sigma}_{f_*\vert f}=E[({\bf f}_*-{\bf m}_{f_*\vert f})({\bf f}_*-{\bf m}_{f_*\vert f})^T]$](img1338.svg) for the variation of
for the variation of  with respect to
with respect to
 , and
, and
![${\bf\Sigma}_{m_{f_*\vert f}}=E_f[({\bf m}_{f_*\vert f}-{\bf m}_{f_*})^2]$](img1333.svg) for the
variation of
for the
variation of
 with respect to
with respect to
 :
:
 given
in the Appendices.
given
in the Appendices.
Now that the posterior



 |
 |
 |
|
 |
![$\displaystyle E_f[ \sigma({\bf f}({\bf X}_*)) ]=\sigma(E_f({\bf f}({\bf X}_*)))
=\sigma( {\bf m}_{f_*} )$](img1348.svg) |
(295) |
 .
.
The Matlab code for the essential parts of this algorithm is
listed below. Here X and y are for the training data

Xs is an array composed
of test vectors. First, the essential segment of the main program
listed below takes in the training and test data, generates the
covariance matrices 


K, Ks, and Kss, respectively.
The function Kernel is exactly the same as the one used for
Gaussian process regression. This code segment then further calls
a function findPosteriorMean which finds the mean and
covariance of







K=Kernel(X,X); % cov(f,f), covariance of prior p(f|X)
Ks=Kernel(X,Xs); % cov(f_*,f)
Kss=Kernel(Xs,Xs); % cov(f_*,f_*)
[Sigmaf W w]=findPosteriorMean(K,y);
% get mean/covariance of p(f|D), W, w
meanfD=Ks'*w; % mean of p(f_*|X_*,D)
SigmafD=Kss-Ks'*inv(K-inv(W))*Ks; % covariance of p(f_*|X_*,D)
ys=sign(meanfD); % binary classification of test data
p=1./(1+exp(-meanfD)); % p(y_*=1|X_*,D) as logistic function
The function findPosteriorMean listed below uses Newton's method
to find the mean and covariance of the posterior


meanf and covf, respectively, together with w
and W for the gradient and Hessian of the likelihood



function [meanf covf W w]=findPosteriorMean(K,y)
% K: covariance of prior of p(f|X)
% y: labeling of training data X
% w: gradient vector of log p(y|f)
% W: Hessian matrix of log p(y|f)
% meanf: mean of p(f|X,y)
% covf: covariance of p(f|X,y)
n=length(y); % number of training samples
f0=zeros(n,1); % initial value of latent function
f=f0;
er=1;
while er > 10^(-9) % Newton's method to get f that maximizes p(f|X,y)
e=exp(-y.*f);
w=y.*e./(1+e); % update w
W=diag(-e./(1+e).^2); % update W
f=inv(inv(K)-W)*(w-W*f0); % iteration to get f from previous f0
er=norm(f-f0); % difference between two consecutive f's
f0=f; % update f
end
meanf=f; % mean of f
covf=inv(inv(K)-W); % coviance of f
end
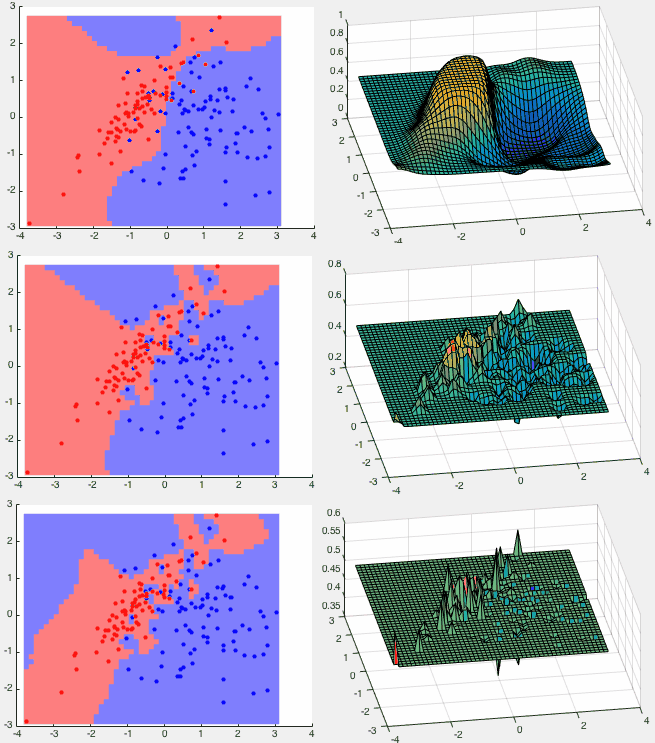
Example 1 The GPC method is trained by the two classes shown in
the figure below, represented by 100 red points and 80 blue points
drawn from two Gaussian distributions


![$\displaystyle {\bf m}_0=\left[\begin{array}{c}1\\ 0\end{array}\right],\;\;\;
{\...
...ht],\;\;\;
{\bf\Sigma}_1=\left[\begin{array}{cc}1&0.9\\ 0.9&1\end{array}\right]$](img1360.svg) |
(296) |
Three classification results are shown in the figure below corresponding
to different values used in the kernel function. On the left, the 2-D
space is partitioned into red and blue regions corresponding to the
two classes based on the sign function of



We see that wherever there is evidence represented by the training
samples of either class in red or blue, there are high probabilities
for the neighboring points to belong to either 

We make the following observations for three different values of the
parameter 
 (top row), the space is partitioned into three regions,
with 20 out of 180 training points misclassified. The estimated
distribution is smooth.
(top row), the space is partitioned into three regions,
with 20 out of 180 training points misclassified. The estimated
distribution is smooth.
 (middle row), the space is fragmented into several
more pieces for the two classes (blue islands inside the red region
and vice versa), with 5 out of the 180 training points misclassified.
The estimated distribution is jagged.
(middle row), the space is fragmented into several
more pieces for the two classes (blue islands inside the red region
and vice versa), with 5 out of the 180 training points misclassified.
The estimated distribution is jagged.
 (bottom row), the space is partitioned into still
more pieces, with all 180 training points correctly classified. The
estimated distribution is spiky.
(bottom row), the space is partitioned into still
more pieces, with all 180 training points correctly classified. The
estimated distribution is spiky.
 is small, the classificatioin
result is not necessarily the best as it may well be an overfitting of
the noisy data. We conclude that by adjusting parameter
is small, the classificatioin
result is not necessarily the best as it may well be an overfitting of
the noisy data. We conclude that by adjusting parameter  , we can
make proper tradeoff between error rate and overfitting.
, we can
make proper tradeoff between error rate and overfitting.