Next: Multi-Class Classification Up: Support Vector machine Previous: Soft Margin SVM
The sequential minimal optimization (SMO, due to John Platt 1998, also see notes here) is a more efficient algorithm for solving the SVM problem, compared with the generic QP algorithms such as the internal-point method. The SMO algorithm can be considered as a method of decomposition, by which an optimization problem of multiple variables is decomposed into a series of subproblems each optimizing an objective function of a small number of variables, typically only one, while all other variables are treated as constants that remain unchanged in the subproblem. For example, the coordinate descent algorithm is just such a decomposition method, which solves a problem in a multi-dimensional space by converting it into a sequence of subproblems each in a one-dimensional space.
When applying the decomposition method to the soft margin SVM problem,
we could consider optimizing only one variable 









 are replaced by the corresponding kernel
function
are replaced by the corresponding kernel
function
 with
with
 .
.
This maximization problem can be solved iteratively. Out of all
previous values




We rewrite the second constraint as
 |
(136) |
 to get
where we have defined
to get
where we have defined  and
and
 . As
. As  is
independent of
is
independent of  and
and  , it remains the
same before and after they are updated, and we have
, it remains the
same before and after they are updated, and we have
 |
(138) |
We now consider a closed-form solution for updating 

 |
(140) |
 , now denoted by
, now denoted by
 , can be written as
, can be written as
 |
 |
 |
|
 |
 |
||
 |
 |
(141) |
 |
(142) |
 . Now the objective function above can be written as:
. Now the objective function above can be written as:
 |
 |
 |
(143) |
 (Eq. (139))
into
(Eq. (139))
into
 , we can rewrite it as a function of
a single variable
, we can rewrite it as a function of
a single variable  alone:
alone:
 |
 |
![$\displaystyle \delta+(1-s)\alpha_j
-\frac{1}{2}\left[ (\delta-s\alpha_j)^2K_{ii}
+\alpha_j^2K_{jj}+2s(\delta-s\alpha_j)\alpha_jK_{ij}\right]$](img504.svg) |
|
 |
|||
 |
![$\displaystyle \frac{1}{2}(2K_{ij}-K_{ii}-K_{jj})\alpha_j^2
+[1-s+s\delta(K_{ii}-K_{ij})+y_j(v_i-v_j)]\alpha_j+$](img506.svg) Const. Const. |
(144) |
 , and scalar Const. represents all constant
terms independent of
, and scalar Const. represents all constant
terms independent of  and
and  and can therefore be
dropped. Now the objective function becomes a quadratic function of
the single variable
and can therefore be
dropped. Now the objective function becomes a quadratic function of
the single variable  . Consider the coefficients of this
quadratic function:
. Consider the coefficients of this
quadratic function:
 :
:
 |
(145) |
 is defined as
is defined as
 |
 |
 |
|
 |
 |
(146) |
 :
:
 |
|||
 |
 |
||
![$\displaystyle +y_j[(u_i-b-\alpha_iy_iK_{ii}-\alpha_jy_jK_{ij})-
(u_j-b-\alpha_iy_iK_{ij}-\alpha_jy_jK_{jj})]$](img516.svg) |
|||
 |
 |
||
 |
|||
 |
 |
||
 |
 |
||
 |
 |
(147) |
 |
(148) |
 and the
actual output based on the previous values of the variables
and the
actual output based on the previous values of the variables
 .
.
![$\displaystyle L(\alpha_j)=\frac{1}{2}\eta\alpha_j^2
+\left[y_j(E_i-E_j)-\alpha_j^{old}\eta\right]\alpha_j$](img525.svg) |
(149) |
 in
the coefficients, we will find
in
the coefficients, we will find
 that maximizes
that maximizes
 . Consider its first and second order derivatives:
. Consider its first and second order derivatives:
 |
 |
 |
|
 |
 |
 |
(150) |
 is
is  , it
has a maximum. Solving the equation
, it
has a maximum. Solving the equation
 , we
get
, we
get
 that maximizes
that maximizes
 based on the old
value
based on the old
value
 :
:
 |
(151) |
Note that this is an unconstrained solution, which needs to be modified
if the constraints in Eq. (135) are violated. As shown in
the figure below, 




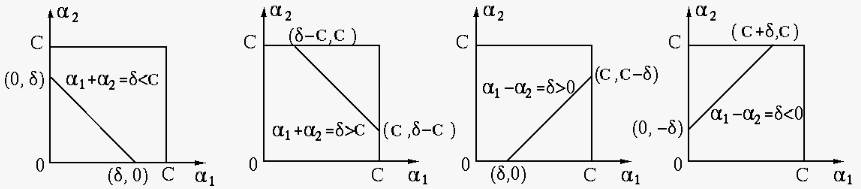
The four panels correspond to the four possible cases, depending
on whether 








 , then
, then
 ,
,
 , then
, then
 (first panel)
(first panel)
 , then
, then
 (second panel)
(second panel)
 |
(152) |
 , then
, then
 ,
,
 , then
, then
 (third panel)
(third panel)
 , then
, then
 (fourth panel)
(fourth panel)
 |
(153) |
Now we apply the constraints in terms of 



 |
(154) |
 based on Eq. (139):
based on Eq. (139):
 |
(155) |
 |
 |
 |
|
 |
 |
||
 |
 |
(156) |
 .
To update
.
To update  , consider:
, consider:
 |
 |
 |
|
 |
 |
||
 |
 |
(157) |
 for either
for either  or
or  , then
, then  according to Eq. (130). We let
according to Eq. (130). We let
 and
solve the equation above to get:
and
solve the equation above to get:
 or or |
(158) |
 for both
for both  and
and  , then
, then
 .
.
 or
or
 for both
for both  and
and  ,
then
,
then  , and
, and
 , their average
, their average
 can be used.
can be used.
The computation above for maximizing










The Matlab code for the essential part of the SMO algorithm is listed below, based mostly on a simplified SMO algorithm and the related code with some modifications.
In particular, the if-statement
if (alpha(i)<C & yE<-tol) | (alpha(i)>0 & yE>tol)checks the three KKT conditions in Eq. (130), where
 and
and  is a small constant. The first half checks if
the first two of the three conditions are violated, while the second
half checks if the second two of the three conditions are violated.
is a small constant. The first half checks if
the first two of the three conditions are violated, while the second
half checks if the second two of the three conditions are violated.
[X y]=getData; % get training data
[m n]=size(X); % size of data
alpha=zeros(1,n); % alpha variables
bias=0; % initial bias
it=0; % iteration index
while (it<maxit) % number of iterations less than maximum
it=it+1;
changed_alphas=0; % number of changed alphas
N=length(y); % number of support vectors
for i=1:N % for each alpha_i
Ei=sum(alpha.*y.*K(X,X(:,i)))+bias-y(i);
yE=Ei*y(i);
if (alpha(i)<C & yE<-tol) | (alpha(i)>0 & yE>tol) % KKT violation
for j=[1:i-1,i+1:N] % for each alpha_j not equal alpha_i
Ej=sum(alpha.*y.*K(X,X(:,j)))+bias-y(j);
ai=alpha(i); % alpha_i old
aj=alpha(j); % alpha_j old
if y(i)==y(j) % s=y_i y_j=1
L=max(0,alpha(i)+alpha(j)-C);
H=min(C,alpha(i)+alpha(j));
else % s=y_i y_j=-1
L=max(0,alpha(j)-alpha(i));
H=min(C,C+alpha(j)-alpha(i));
end
if L==H % skip when L==H
continue
end
eta=2*K(X(:,j),X(:,i))-K(X(:,i),X(:,i))-K(X(:,j),X(:,j));
alpha(j)=alpha(j)+y(j)*(Ej-Ei)/eta; % update alpha_j
if alpha(j) > H
alpha(j) = H;
elseif alpha(j) < L
alpha(j) = L;
end
if norm(alpha(j)-aj) < tol % skip if no change
continue
end
alpha(i)=alpha(i)-y(i)*y(j)*(alpha(j)-aj); % find alpha_i
bi = bias - Ei - y(i)*(alpha(i)-ai)*K(X(:,i),X(:,i))...
-y(j)*(alpha(j)-aj)*K(X(:,j),X(:,i));
bj = bias - Ej - y(i)*(alpha(i)-ai)*K(X(:,i),X(:,j))...
-y(j)*(alpha(j)-aj)*K(X(:,j),X(:,j));
if 0<alpha(i) & alpha(i)<C
bias=bi;
elseif 0<alpha(j) & alpha(j)<C
bias=bj;
else
bias=(bi+bj)/2;
end
changed_alphas=changed_alphas+1; % one more alpha changed
end
end
end
if changed_alphas==0 % no more changed alpha, quit
break
end
I=find(alpha~=0); % indecies of non-zero alphas
alpha=alpha(I); % find non-zero alphas
Xsv=X(:,I); % find support vectors
ysv=y(I); % their corresponding indecies
end % end of iteration
nsv=length(ysv); % number of support vectors
where K(X,x) is a function that takes an  matrix
matrix
![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_n]$](img585.svg) and an n-D vector
and an n-D vector  and produces the kernel functions
and produces the kernel functions
 as output.
as output.
function ker=K(X,x)
global kfunction
global gm % parameter for Gaussian kernel
[m n]=size(X);
ker=zeros(1,n);
if kfunction=='l'
for i=1:n
ker(i)=X(:,i).'*x; % linear kernel
end
elseif kfunction=='g'
for i=1:n
ker(i)=exp(-gm*norm(X(:,i)-x)); % Gaussian kernel
end
elseif kfunction=='p'
for i=1:n
ker(i)=(X(:,i).'*x).^3; % polynomial kernel
end
end
end
Having found all non-zero
 and the corresponding
support vectors, we further find the offset or bias term:
and the corresponding
support vectors, we further find the offset or bias term:
bias=0;
for i=1:nsv
bias=bias+(ysv(i)-sum(ysv.*alpha.*K(Xsv,Xsv(:,i))));
end
bias=bias/nsv;
Any unlabeled data point  is classified into class
is classified into class  or
or
 , depending on whether the following expression is greater or
smaller than zero:
, depending on whether the following expression is greater or
smaller than zero:
y=sum(alpha.*ysv.*K(Xsv,x))+bias;
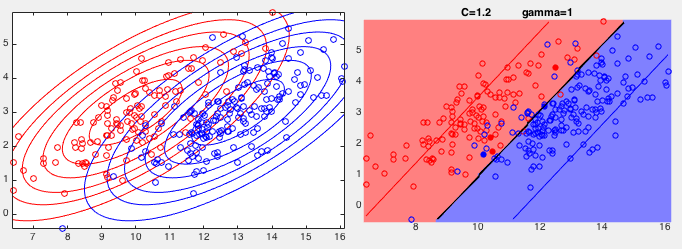
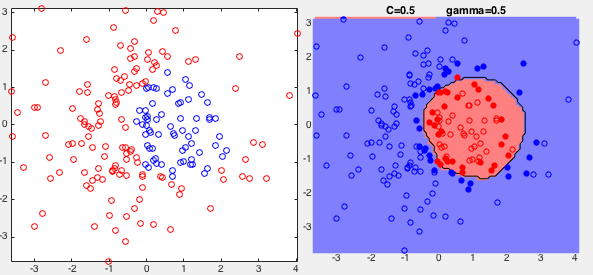
Example 1: The same training set used previously to test the SVM
with a hard margin is used again to test the SVM with a soft margin.
Two results are shown below, corresponding to two different values





On the other hand, when 

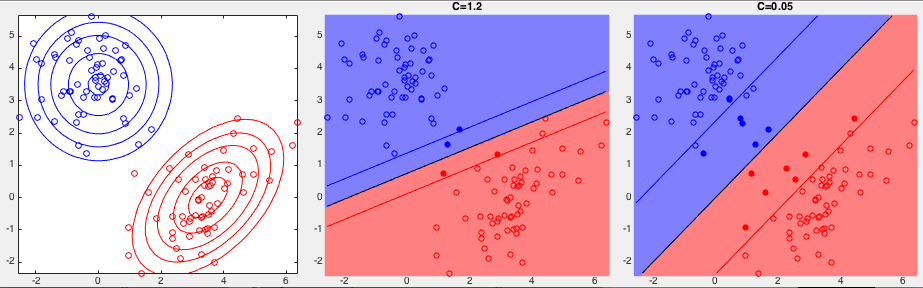
Example 2: The classification results for the XOR data set
are shown below with 



One the other hand, when 
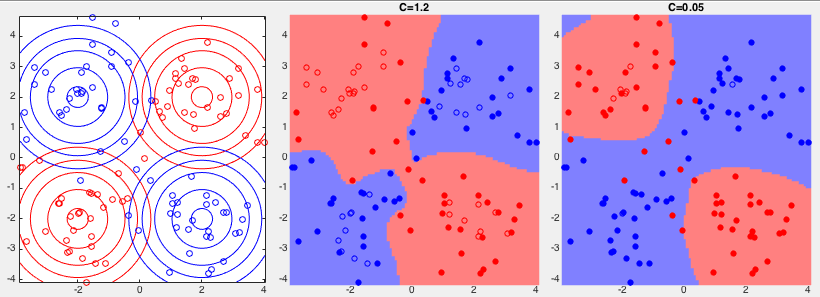
Example 3:
Example 4: