Next: Kernel Mapping Up: Support Vector machine Previous: Support Vector machine
The support vector machine (SVM) is a supervised binary classifier
trained by a dsta set







 |
(62) |
 and intercept
and intercept  . Once
the two parameters
. Once
the two parameters  and
and  are determined based on the
training set, any unlabeled sample
are determined based on the
training set, any unlabeled sample  can be classified into
either of the two classes:
can be classified into
either of the two classes:
if then then |
(63) |
The initial setup of the SVM algorithm seems the same as the
method of linear regression
as a linear binary classifier when the linear regression function




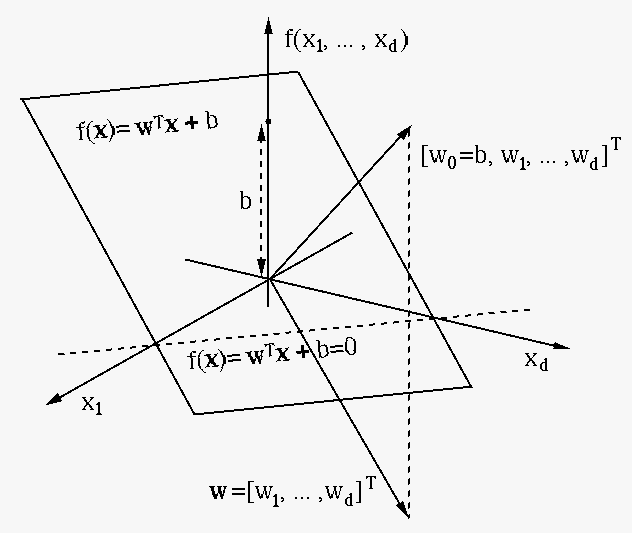
We rewrite the decision equation

 |
(64) |
 on the decision plane
on the decision plane  onto its normal direction
onto its normal direction  ,
of which the absolute value is the distance between
,
of which the absolute value is the distance between  and the
origin:
We further find the projection of any point
and the
origin:
We further find the projection of any point
 off the decision plane
off the decision plane  onto
onto  as
as
 , and its distance
to
, and its distance
to  as:
as:
 |
(66) |
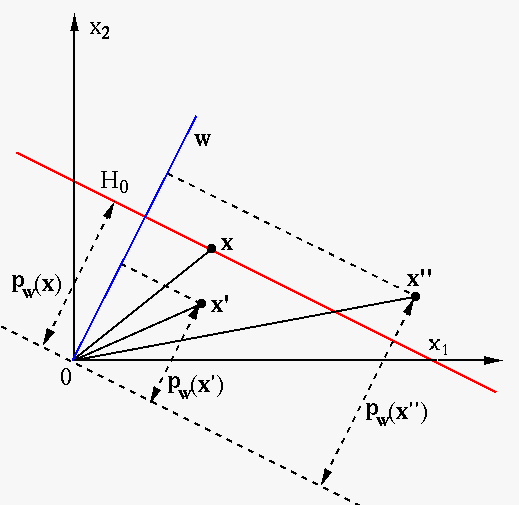
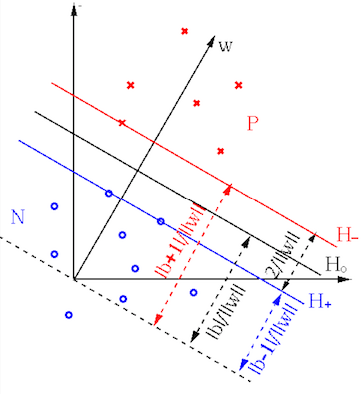
We desire to find the optimal decision plane 

 |
(67) |
 and
and  that
are in parallel with
that
are in parallel with  and pass through the support vectors
and pass through the support vectors
 on either side of
on either side of  :
As these equations can be arbitrarily scaled, we can let
:
As these equations can be arbitrarily scaled, we can let  for
convenience. Based on Eq. (65), the distances from
for
convenience. Based on Eq. (65), the distances from
 or
or  to the origin can be written as
to the origin can be written as
 |
(69) |
 , called the decision
margin, can be found as:
To maximize this margin, we need to minimize
, called the decision
margin, can be found as:
To maximize this margin, we need to minimize
 .
.
For these planes to correctly separate all samples in the training set

 |
(71) |
 or
or
 , while the inequalities are satisfied by all other samples
farther away from
, while the inequalities are satisfied by all other samples
farther away from  behind
behind  or
or  . The two conditions
above can be combined to become:
Now the task of finding the optimal decision plane
. The two conditions
above can be combined to become:
Now the task of finding the optimal decision plane  can be
formulated as a constrained minimization problem:
can be
formulated as a constrained minimization problem:
Example:
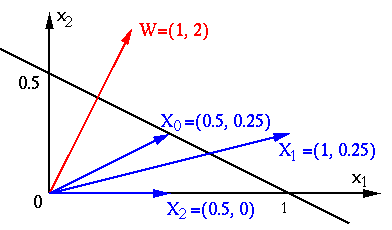
The straight line in 2D space shown above, denoted by 
![$\displaystyle f({\bf x})={\bf w}^T{\bf x}+b=[w_1,w_2]
\left[ \begin{array}{c} x...
...b
=[1, 2]\left[ \begin{array}{c} x_1 \\ x_2 \end{array} \right]-1
=x_1+2x_2-1=0$](img290.svg) |
(74) |
 to the origin is:
to the origin is:
 |
(75) |
![${\bf x}_0=[0.5,\;0.25]^T$](img292.svg) ,
,
 ,
i.e.,
,
i.e.,  is on the plane. Its distance to the plane is
is on the plane. Its distance to the plane is
 .
.
![${\bf x}_1=[1,\;0.25]^T$](img296.svg) ,
,
 ,
i.e.,
,
i.e.,  is above the straight line, its distance to the plane
is
is above the straight line, its distance to the plane
is
 .
.
![${\bf x}_2=[0.5,\;0]^T$](img300.svg) ,
,
 , i.e.,
, i.e.,
 is below the straight line, its distance to the plane is
is below the straight line, its distance to the plane is
 .
.
These two points
![${\bf x}_1=[1,\,0.25]^T$](img304.svg)
![${\bf x}_2=[0.5,\,0]^T$](img305.svg)




 |
(76) |
 on both sides of the decision equation
on both sides of the decision equation
 ,
it is scaled to become
,
it is scaled to become
 |
(77) |
 parallel to
parallel to  and passing
through
and passing
through  and
and  passing through
passing through  are
are
 |
 |
||
 |
 |
 |
(78) |
 and
and  is indeed
is indeed
 :
:
 |
(79) |
 and the origin
and the origin
 found previously.
found previously.
For reasons to be discussed later, instead of directly solving the constrained minimization problem in Eq. (73), now called the primal problem, we actually solve the dual problem. Specifically, we first construct the Lagrangian function of the primal problem:
where are the Lagrange multipliers,
which is called the primal function. Here for this minimization
problem with non-positive constraints, the Lagrangian multipliers are
required to be negative,
are the Lagrange multipliers,
which is called the primal function. Here for this minimization
problem with non-positive constraints, the Lagrangian multipliers are
required to be negative,  , according to Table
, according to Table ![[*]](crossref.png) here, if a minus sign is used
for the second term. However, to be consistent with most SVM literatures,
we use the positive sign for the second term and require
here, if a minus sign is used
for the second term. However, to be consistent with most SVM literatures,
we use the positive sign for the second term and require
 .
Note that if
.
Note that if  is a support vector on either
is a support vector on either  or
or  ,
i.e., the equality constraint
,
i.e., the equality constraint
 holds, then
holds, then
 ; but if
; but if  is not a support vector, the equality
constraint does not hold, and
is not a support vector, the equality
constraint does not hold, and
 .
.
We next find the minimum (or infimum) as the lower bound of the primal
function



![$\displaystyle \frac{\partial}{\partial {\bf w}}L_p({\bf w},b)
=\frac{\partial}{...
...\bf w}^T {\bf x}_n+b))\right]
={\bf w}-\sum_{n=1}^N\alpha_ny_n{\bf x}_n={\bf0},$](img327.svg) |
(82) |
 , we get its lower bound as a function
of the Lagrange multipliers
, we get its lower bound as a function
of the Lagrange multipliers
 , called
the dual function:
, called
the dual function:
 |
 |
![$\displaystyle \inf_{{\bf w},b} L_p({\bf w},b,{\bf\alpha})
=\inf_{{\bf w},b}\lef...
...}{\bf w}^T{\bf w}
+\sum_{n=1}^N \alpha_n(1-y_n( {\bf w}^T {\bf x}_n+b)) \right]$](img332.svg) |
|
 |
![$\displaystyle \inf_{{\bf w},b}\left[\frac{1}{2}{\bf w}^T{\bf w}
+\sum_{n=1}^N \...
...\bf w}^T \sum_{n=1}^N \alpha_ny_n{\bf x}_n
-b\;\sum_{n=1}^N \alpha_ny_n \right]$](img333.svg) |
||
 |
 |
||
 |
 |
(84) |
 with respect to the Lagrange multipliers, subject to the constraint
imposed by Eq. (81):
with respect to the Lagrange multipliers, subject to the constraint
imposed by Eq. (81):
 is an
is an  by
by  symmetric matrix of which the component
in the mth row and nth column is
symmetric matrix of which the component
in the mth row and nth column is
 (
(
 ). Now we have converted the primal problem of
constrained minimization of the primal function
). Now we have converted the primal problem of
constrained minimization of the primal function
 with respect to
with respect to  and
and  to its dual problem of linearly
constrained maximization of the dual function
to its dual problem of linearly
constrained maximization of the dual function
 with respect to
with respect to
 . Solving this
quadratic programming (QP) problem,
we get all Lagrange multipliers
. Solving this
quadratic programming (QP) problem,
we get all Lagrange multipliers
 .
All training samples
.
All training samples  corresponding to positive Lagrange
multipliers
corresponding to positive Lagrange
multipliers
 are support vectors, while others corresponding
to
are support vectors, while others corresponding
to
 are not support vectors.
are not support vectors.
We can now find the normal direction 
 is the difference between the
weighted means of the support vectors in class
is the difference between the
weighted means of the support vectors in class  (first term)
and class
(first term)
and class  (second term), and it is determined only by the
support vectors.
(second term), and it is determined only by the
support vectors.
Having found


 or or |
(87) |
 ). Solving the equation for
). Solving the equation for  we get:
All support vectors should yield the same result. Computationally we
simply get
we get:
All support vectors should yield the same result. Computationally we
simply get  as the average of the above for all support vectors.
as the average of the above for all support vectors.
We note that both 








Having obtained 


 |
 |
 |
|
 |
 |
(90) |
 . Now the decision margin can be written as:
. Now the decision margin can be written as:
 |
(91) |
The SVM algorithm for binary classification can now by summarized as the following steps:
 by solving the QP problem in Eq. (85);
by solving the QP problem in Eq. (85);
 by Eq. (86);
by Eq. (86);
 by Eq. (88);
by Eq. (88);
 by Eq. (89).
by Eq. (89).
 as well as
as well as
 and
and  in the training set, the normal vector
in the training set, the normal vector  is never needed, and the second step above for calculating
is never needed, and the second step above for calculating  by Eq. (86) can be dropped.
by Eq. (86) can be dropped.
We prefer to solve this dual problem not only because it is easier
than the original primal problem, but also, more importantly, for
the reason that all data points appear only in the form of an inner
product

The Matlab code for the essential part of the algorithm is listed
below, where X and y are respectively the data array
composed of 
![$[{\bf x}_1,\cdots,{\bf x}_N]$](img356.svg)

IPmethod
is a function that implements the
interior point method
for solving a general QP problem of minimizing



[X y]=getData; % get the training data
[m n]=size(X);
Q=(y'*y).*(X'*X); % compute the quadratic matrix
c=-ones(n,1); % coefficients of linear term
A=y; % coefficient matrix and
b=0; % constants of linear equality constraints
alpha=0.1*ones(n,1); % initial guess of solution
[alpha mu lambda]=IPmethod(Q,A,c,b,alpha); % solve QP to find alphas
% by interior point method
I=find(abs(alpha)>10^(-3)); % indecies of non-zero alphas
asv=alpha(I); % non-zero alphas
Xsv=X(:,I); % support vectors
ysv=y(:,I); % their labels
w=sum(repmat(ysv.*asv,m,1).*Xsv,2); % normal vector (not needed)
bias=mean(ysv-w'*Xsv); % bias
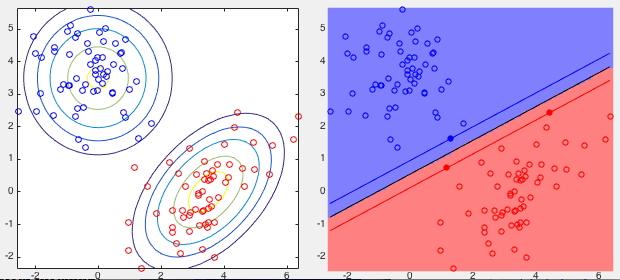
Example: The training set contains two classess of 2-D points with Gaussian distributions generated based on the following mean vectors and covariance matrices:
![$\displaystyle {\bf m}_-=\left[\begin{array}{c}3.5\\ 0\end{array}\right],\;\;\;\...
...y}\right],\;\;\;\;
{\bf S}_+=\left[\begin{array}{cc}1&0\\ 0&1\end{array}\right]$](img361.svg) |
(92) |
 and constant
and constant  of the optimal boundary between the two classes based on three support
vectors, all listed below. Note that decision boundary is completely
dictated by the three support vectors and the margion distance between
them is maximized.
of the optimal boundary between the two classes based on three support
vectors, all listed below. Note that decision boundary is completely
dictated by the three support vectors and the margion distance between
them is maximized.
![\begin{displaymath}\begin{array}{c\vert c\vert l\vert r}\hline
n & \alpha_n & {\...
...rray}{r}-1.25\\ 2.39\end{array}\right],\;\;\;\;\;\;\;\;
b=-1.31\end{displaymath}](img362.svg) |
(93) |