Next: Naive Bayes Classification Up: ch9 Previous: Discriminative vs. Generative Methods
Here we first consider a set of simple supervised classification
algorithms that assign an unlabeled sample 


![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img9.svg)



Given an unlabeled pattern 






While the k-NN method is simple and straight forward, its computational
cost is high as classifying any unlabeled pattern 
Given a set of training data points



 |
(1) |
 can be classified to one of the
can be classified to one of the
 classes based on certain distance
classes based on certain distance
 between
between
 and each of class
and each of class  :
:
if then then |
(2) |
We could simply use the Euclidean distance





Example 1: As illustrated in the figure below (left plot), a
point 
 |
(3) |
 and
and  are considered, we have
are considered, we have
 , i.e.,
, i.e.,  is closer to
is closer to  than
than  and therefore should be classified to class
and therefore should be classified to class  . However,
as shown in the plot,
. However,
as shown in the plot,  should be classified to class
should be classified to class  , if the
variances
, if the
variances
 and
and
 are also taken into consideration.
are also taken into consideration.
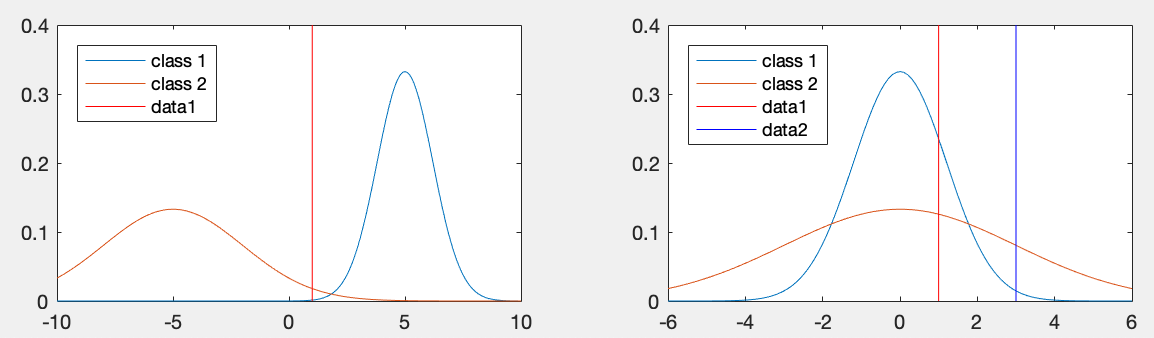
We see the distance







In a higher dimensional feature space, we can carry out classification
based on the more generally defined Mahalanobis distance between
a point 


 |
(4) |
Example 2: As illustrated in the above figure (right plot), two
samples 

 |
(5) |
 are the same,
are the same,
 for both samples
for both samples  and
and  , they are both classified into
, they are both classified into  with a greater variance
with a greater variance
 therefore smaller
Mahalanobis distances:
therefore smaller
Mahalanobis distances:
 |
(6) |
 should be classified to
should be classified to  .
We therefore see that sometimes the Mahalanobis distance is not reliable
for classification, and some better method need to be considered, as
discussed later.
.
We therefore see that sometimes the Mahalanobis distance is not reliable
for classification, and some better method need to be considered, as
discussed later.