Next: t-Distributed Stochastic Neighbor Embedding Up: ch8 Previous: Probabilistic PCA
The goal of multidimensional scaling (MDS) is to reduce
the dimensionality of the dataset representing a set of 

![${\bf x}=[x_1,\cdots,x_d]^T$](img2.svg)
Specifically, we represent the pairwise similarity of a set of 
![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img6.svg)

![${\bf D}_x=[\;d^x_{ij}\;]$](img621.svg)




![${\bf Y}=[{\bf y}_1,\cdots,{\bf y}_N]$](img625.svg)

![${\bf D}_y=[\; d^y_{ij} ]$](img627.svg)






We assume the given pairwise similarity is the Euclidean distance



 |
(170) |
![$\displaystyle {\bf D}_x^2=\left[\begin{array}{ccc} & & \\
& d^2_{ij} & \\ & & ...
... &\\
& & \end{array}\right]_{N\times N}
={\bf X}_r-2{\bf X}^T{\bf X}+{\bf X}_c$](img632.svg) |
(171) |
 |
 |
![$\displaystyle \left[\begin{array}{ccc}\vert\vert{\bf x}_1\vert\vert^2&\cdots&\v...
...ert\vert^2&\cdots&\vert\vert{\bf x}_N\vert\vert^2\end{array}\right]={\bf X}_c^T$](img634.svg) |
|
 |
 |
![$\displaystyle \left[\begin{array}{c}{\bf x_1}^T\\ \vdots\\ {\bf x}_N^T\end{arra...
...ts&\vdots\\
{\bf x}_N^T{\bf x}_1&\cdots&{\bf x}_N^T{\bf x}_N\end{array}\right]$](img636.svg) |
(172) |
Our goal is to find
![${\bf Y}=[{\bf y}_1,\cdots,{\bf y}_N]$](img625.svg)


 |
(173) |
We note that such a solution 



 |
(174) |
To do so, we introduce an 




![${\bf a}=[a_1,\cdots,a_N]^T$](img643.svg)


![$\displaystyle {\bf Ca} ={\bf Ia}-\frac{1}{N}{\bf 1}{\bf a}
=\left[\begin{array}...
...t]
=\left[\begin{array}{c}a_1-\bar{a}\\ \vdots\\ a_N-\bar{a}
\end{array}\right]$](img645.svg) |
(175) |
![$\displaystyle ({\bf Ca})^T={\bf a}^T{\bf C}=[a_1-\bar{a},\cdots,a_N-\bar{a}]$](img646.svg) |
(176) |
 to a row vector
to a row vector
![${\bf a}^T=[a_1,\cdots,a_N]$](img647.svg) removes the mean
removes the mean  from
each of its component.
from
each of its component.
We can now postmultiply 


![$\displaystyle {\bf XC}=[{\bf x}_1,\cdots,{\bf x}_N]{\bf C}
=[\bar{\bf x}_1,\cdots,\bar{\bf x}_N]=\bar{\bf X}$](img649.svg) |
(177) |
 where where |
(178) |
 to
to  to remove the mean of
each column of
to remove the mean of
each column of  :
:
 |
(179) |
We further apply double centering to


 |
 |
 |
|
 |
 |
 ,
as all components of each row of
,
as all components of each row of  are the same as
their mean, and removing the mean results in a zero row vector,
are the same as
their mean, and removing the mean results in a zero row vector,
 ; and the last term
; and the last term
 ,
as all components of each column of
,
as all components of each column of  are the same
as their mean, and removing the mean results in a zero column
vector.
are the same
as their mean, and removing the mean results in a zero column
vector.
Now the similarity matrix



 |
(180) |
We further assume all data points in 

 |
(181) |
 that minimizes
that minimizes
 , we first consider
the equation
, we first consider
the equation
 so
that
so
that
 . In other words, we desire to express
. In other words, we desire to express
 as the inner product of some matrix with itself.
To do so, we carry out eigenvalue decomposition of
as the inner product of some matrix with itself.
To do so, we carry out eigenvalue decomposition of  to
find its eigenvalue matrix
to
find its eigenvalue matrix
 and the orthogonal
eigenvector matrix
and the orthogonal
eigenvector matrix  satisfying
satisfying
 , i.e.,
, i.e.,
 |
 |
![$\displaystyle {\bf V}{\bf\Lambda}{\bf V}^{-1}
={\bf V}{\bf\Lambda}{\bf V}^T=[{\...
...ght]
\left[\begin{array}{c}{\bf v}_1^T\\ \vdots\\ {\bf v}_N^T\end{array}\right]$](img674.svg) |
|
 |
 |
 is symmetric, the eigenvalues are real
and the eigenvectors are orthogonal, i.e.,
is symmetric, the eigenvalues are real
and the eigenvectors are orthogonal, i.e.,
 is
is orthogonal matrix. Now we can find the desired
is
is orthogonal matrix. Now we can find the desired  as:
as:
![$\displaystyle {\bf Y}={\bf\Lambda}^{1/2}{\bf V}^T
=\left[\begin{array}{ccc}\sqr...
...lambda_1}{\bf v}_1^T\\
\vdots\\ \sqrt{\lambda_N}{\bf v}_N^T\end{array} \right]$](img677.svg) |
(182) |
 in
in
![${\bf Y}=[{\bf y}_1,\cdots,{\bf y}_N]$](img625.svg) is
an N-D vector. If all
is
an N-D vector. If all  dimensions of these
dimensions of these  are used,
are used,
 and
and
 . To reduce the
dimensionality to
. To reduce the
dimensionality to  , we construct
, we construct  by the first
by the first  rows of
rows of  corresponding to the
corresponding to the  greatest eigenvalues of
greatest eigenvalues of
 and their
corresponding eigenvectors
and their
corresponding eigenvectors
 . The
error is:
. The
error is:
 |
(183) |
In summary, here is the PCoA algorithm:
 matrix
matrix
![${\bf D}_x=[\;d_{ij}\;]$](img686.svg) , construct the squared proximity matrix
, construct the squared proximity matrix
![${\bf D}^2_x=[\;d^2_{ij}\;]$](img687.svg) ;
;
 ;
;
 greatest eigenvalues
greatest eigenvalues
 of
of  , and the corresponding eigenvectors
, and the corresponding eigenvectors
 ;
;
![$\displaystyle {\bf Y}_{d'\times N}={\bf\Lambda}_{d'}^{1/2}{\bf V}_{d'}
=\left[\...
..._1}{\bf v}_1^T\\
\vdots\\ \sqrt{\lambda_{d'}}{\bf v}_{d'}^T\end{array} \right]$](img690.svg) |
(184) |
The Matlab functions for the PCA and PCoA are listed below:
function Y=PCA(X)
[V D]=eig(cov(X')); % solve eigenequation for covariance of data
[d idx]=sort(diag(D),'descend'); % sort eigenvalues in descend order
V=V(:,idx); % reorder eigenvectors
Y=V'*X; % carry out KLT
end
function Y=PCoA(X)
N=size(X,2);
D=zeros(N); % pairwise similarity matrix
C=eye(N)-ones(N)/N; % centering matrix
for i=1:N
for j=1:N
D(i,j)=(norm(X(:,i)-X(:,j)))^2;
end
end
B=-C*D*C/2;
[V D]=eig(B); % solve eigenequation for matrix B
[d idx]=sort(diag(D),'descend'); % sort eigenvalues in descend order
V=V(:,idx); % reorder eigenvectors
Y=sqrt(D)*V'; % carry out PCoA
end
Example 1:
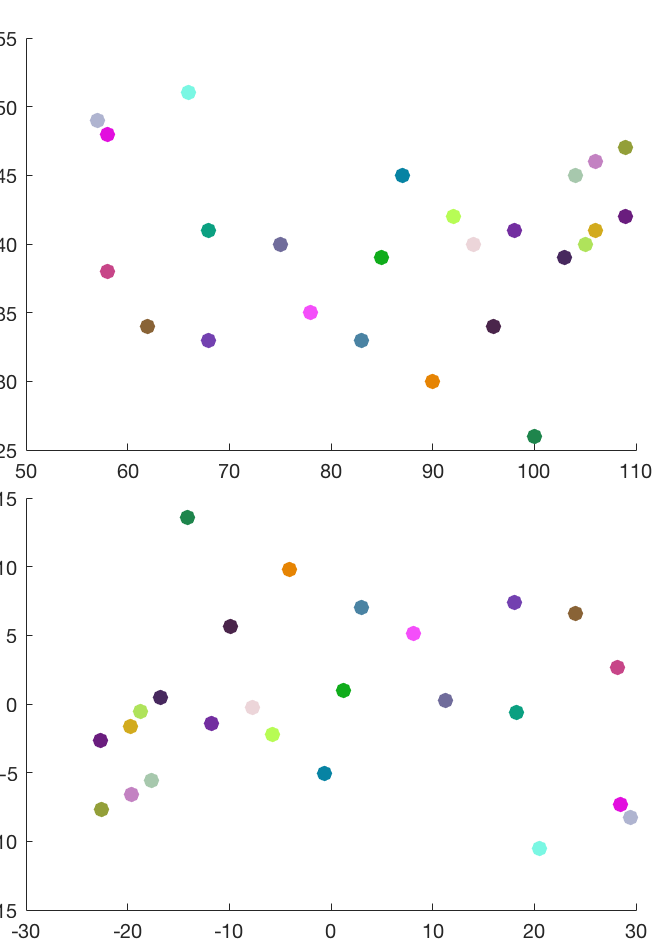
In the figure below, the top panel shows the locations of some major cities in North America, and the bottom panel shows the reconstructed city locations based on the pairwise distance of these cities. Note that the reconstructed map is a rotated and centralized version of the original map.
Example 2:
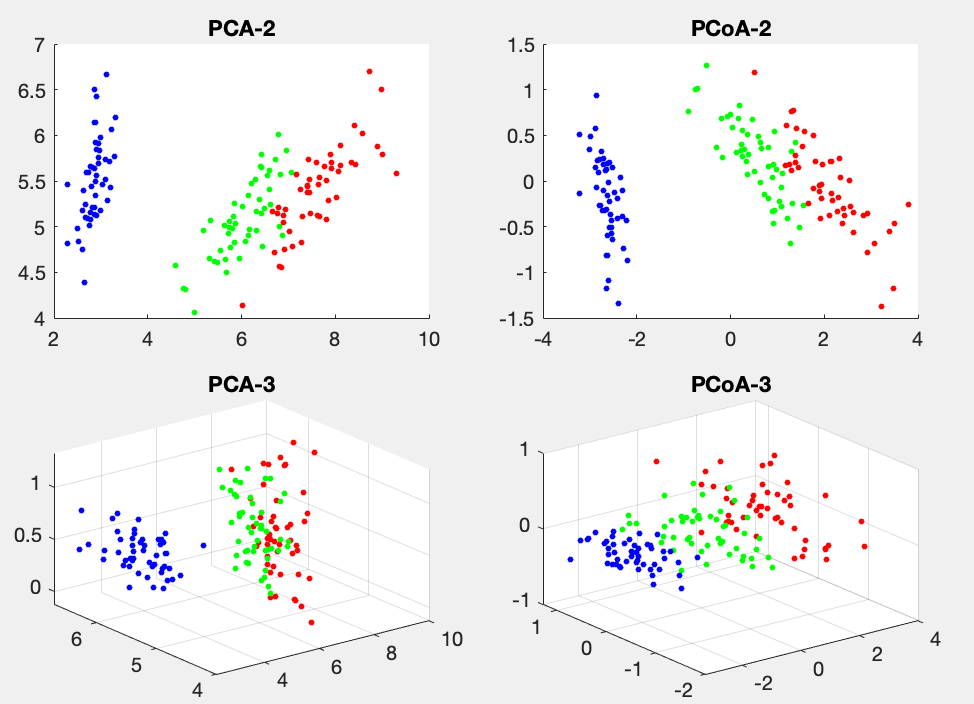
Both PCA and PCoA are applied to the Iris dataset of 150 4-D data points, and the figure below shows the data points in 2-D and 3-D spaces corresponding to the first 2 and 3 eigenvalues.
Example 3:
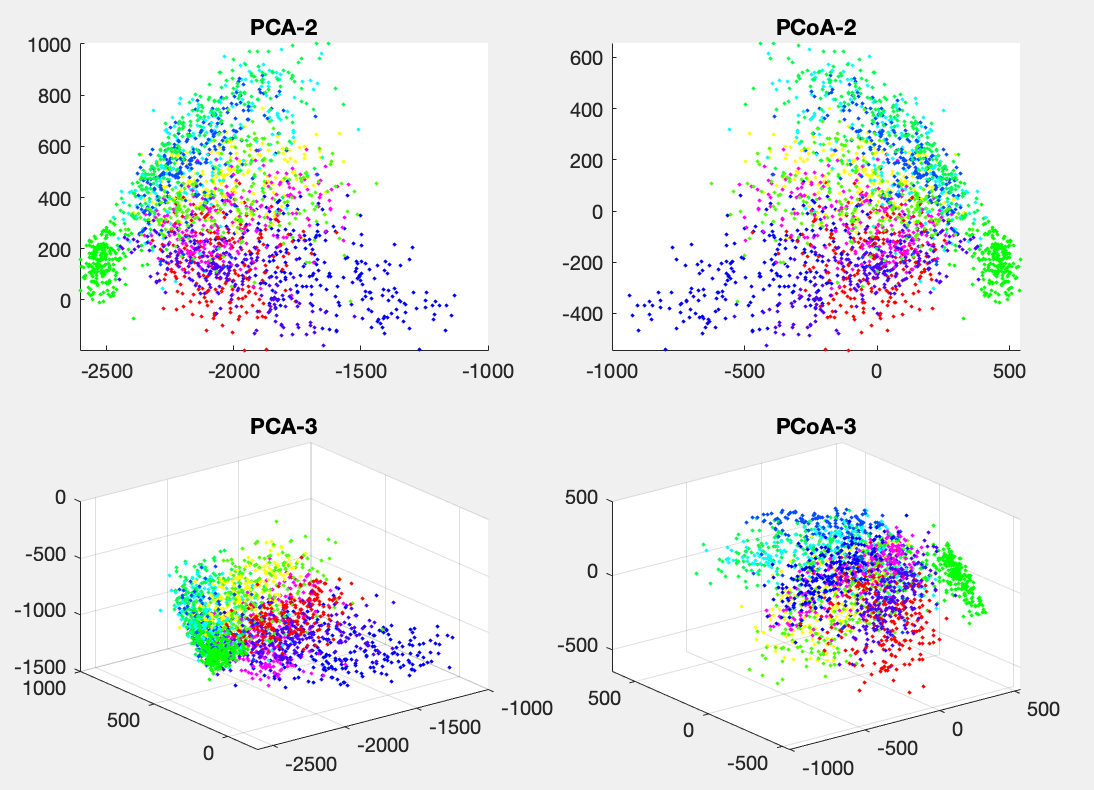
Both PCA and PCoA are applied to the dataset of handwritten numbers of 2240 256-D data points, and the figure below shows the data points in 2-D and 3-D spaces corresponding to the first 2 and 3 eigenvalues.