Next: Independent Component Analysis Up: ch8 Previous: Classical Multidimensional Scaling
It is impossible to reduce the dimensionality of a given dataset which is intrinsically high-dimensional (high-D), while still preserving all the pairwise distances in the resulting low-dimensional (low-D) space, compromise will have to be made to sacrifice certain aspects of the dataset when the dimensionality is reduced.
The PCA method as a linear, dimensionality reduction algorithm,
is unable to interpret complex nonlinear relationship between features. tance preserving method, the PCA emphasizes the importance of the PCA dimensions with great variances, but neglects small distance variations. It places distant and dissimilar data points far apart in the low-D space, while ignoring the importance of similar datapoints close together in the high-D, which need to be also placed close together in the low-D space. Therefore the PCA method tends to preserve large scale global structure of the dataset, while ignoring the small scale local structure.
But in some cases the local structures are of equal or even more importance than the global ones, such as the typical example of “Swiss roll”.
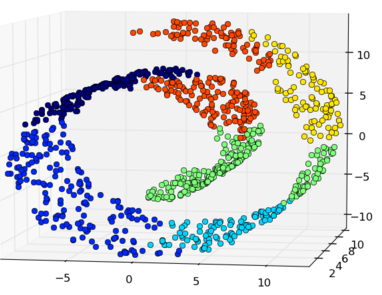
Also, when the dimensionality is much reduced from a high-D space to a low-D space, the crowding problem occurs. In a high-D space, a data point may have a large number of similar neighbors of roughly equal distance; however, in a low-D space, the number of equal-distance neighbors is significantly reduced (8 in a 3-D space, 4 in a 2-D space, 2 in a 1-D space). Thus the space available for the neighbors of a point is much reduced. As the result, when a large number of neighbors in the high-D are mapped to a low-D space, they are forced to spread out, some are getting closer to those that are farther away in the original space, thereby reducing the gaps between potential clusters of similar points.
In contrast, the t-SNE method is a nonlinear method that is based on probability distributions of the data points being neighbors, and it attempts to preserve the structure at all scales, but emphasizing more on the small scale structures, by mapping nearby points in high-D space to nearby points in low-D space.
The similarity between any two distinct points 

 |
(185) |
 ,
,  is defined to be zero, as it
never needed in the algorithm. Note that
is defined to be zero, as it
never needed in the algorithm. Note that  is a probability measurement
as it is normalized and add up to 1.
is a probability measurement
as it is normalized and add up to 1.
If 















Similarly, we could also define the similarity 

 |
(186) |
 be the point of interest (black, in
the center of the Gaussian kernel), then another point
be the point of interest (black, in
the center of the Gaussian kernel), then another point  (red) is
mapped to a different position, which is closer to
(red) is
mapped to a different position, which is closer to  if they are
initially close (similar), or farther away from each other if they are
initially far apart (dissimilar).
if they are
initially close (similar), or farther away from each other if they are
initially far apart (dissimilar).
How well the points 

 |
(187) |
 in the low-D space
that minimize the discrepancy
in the low-D space
that minimize the discrepancy
 .
.
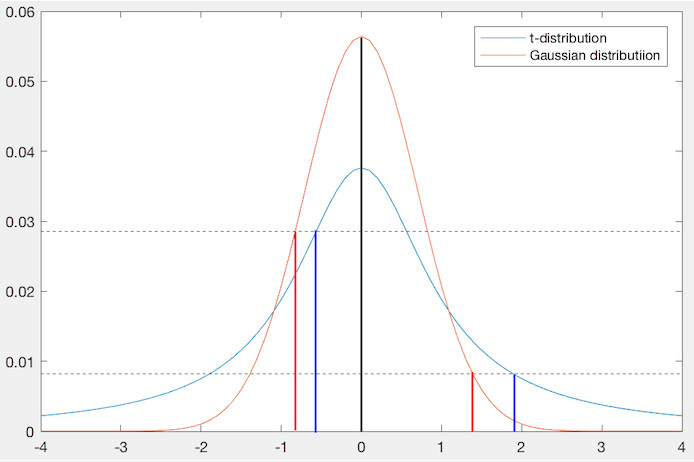
The KL divergence is not symmetric as

 is large but
is large but  is small, the cost
is small, the cost  is very large,
i.e., and such a mapping is heavily penalized. As the result, similar
points that are close to each other in the high-D space tend to be mapped
to points in the low-D space that are also close to each other.
is very large,
i.e., and such a mapping is heavily penalized. As the result, similar
points that are close to each other in the high-D space tend to be mapped
to points in the low-D space that are also close to each other.
 is small but
is small but  is large, the cost
is large, the cost  is small,
i.e., such a mapping is not heavily penalized. As the result, dissimilar
points that are far apart in the high-D space are allowed to be mapped to
points in the low-D space that are closer to each other.
is small,
i.e., such a mapping is not heavily penalized. As the result, dissimilar
points that are far apart in the high-D space are allowed to be mapped to
points in the low-D space that are closer to each other.
Before proceeding to consider the minimization of the cost function 
 defined above is that when a point
defined above is that when a point
 is an outlier with large distances
is an outlier with large distances
 to every other point
to every other point  in the dataset,
in the dataset,  is small and
insignificant in the cost function
is small and
insignificant in the cost function  , and the location of
, and the location of  in the low-D space can not be well determined. To resolve this issue, we
redefine
in the low-D space can not be well determined. To resolve this issue, we
redefine  as the average of two conditioinal probablities:
as the average of two conditioinal probablities:
 |
(188) |
 is the total number of data points and
is the total number of data points and
 |
(189) |
 is a neighboring
point similar to a given point
is a neighboring
point similar to a given point  . Note that
. Note that  is properly
normalized and add up to 1:
is properly
normalized and add up to 1:
 |
(190) |
 for
for  depending on the local data density
in the neighborhood of
depending on the local data density
in the neighborhood of  . Specifically, we choose
. Specifically, we choose  in such
a way that the perplexity
in such
a way that the perplexity
 is the same for all data points, i.e., the entropy
is the same for all data points, i.e., the entropy
 representing the uncertainty is the same,
independent of the local data density. As shown
here, the entropy of a
Gaussian
representing the uncertainty is the same,
independent of the local data density. As shown
here, the entropy of a
Gaussian
 is
is
 , a monotonic function
of its variance
, a monotonic function
of its variance  .
.
We can now find the optimal map points 

To find the gradient of

 |
(191) |
 |
(192) |
![$\displaystyle \frac{\partial Z}{\partial d_{ij}}
=\frac{\partial}{\partial d_{ij}} \left[\sum_k\sum_{l\ne k}(1+d_{kl}^2)^{-1}\right]
=-2d_{ij}(1+d_{ij}^2)^{-2}$](img716.svg) |
(193) |
 based on chain-rule. Note that when
based on chain-rule. Note that when
 changes, only
changes, only  and
and  (for all
(for all  ) change, we therefore
have
) change, we therefore
have
 |
(194) |
![$\displaystyle \frac{\partial d_{ij}}{\partial{\bf y}_i}
=\frac{\partial d_{ji}}...
...f y}_j)\right]^{-1/2}2({\bf y}_i-{\bf y}_j)
=\frac{{\bf y}_i-{\bf y}_j}{d_{ij}}$](img720.svg) |
(195) |
 |
(196) |
 . Note that only
. Note that only  may be a
function of
may be a
function of  , while all
, while all  are constants, we therefore have
are constants, we therefore have
 |
 |
![$\displaystyle \frac{\partial}{\partial d_{ij}}
\left[\sum_k\sum_{l\ne k} \left(...
...right]
=-\sum_k\sum_{l\ne k} p_{kl} \frac{\partial}{\partial d_{ij}}\log q_{kl}$](img726.svg) |
|
 |
 |
||
 |
 |
||
 |
![$\displaystyle 2d_{ij}\left[\frac{p_{ij}}{q_{ij}} \frac{(1+d_{ij}^2)^{-2}}{Z}-\frac{(1+d_{ij}^2)^{-2}}{Z}\right]$](img729.svg) |
(197) |
 when
when  or
or  ,
and the last equality is due to
,
and the last equality is due to
 . But as
. But as
 |
(198) |
 |
(199) |
 with
respect to each of the map points
with
respect to each of the map points  :
:
 |
 |
 |
|
 |
 |
(200) |
Given the gradient vector, we can iteratively approach the optimal data
points in the low-D space that minimize the discrepancy

 |
(201) |
 is the step size (learning rate) and the second term
is momentum term.
is the step size (learning rate) and the second term
is momentum term.