Next: Support Vector machine Up: ch9 Previous: Naive Bayes Classification
The Adaptive boosting (AdaBoost) is a supervised binary
classification algorithm based on a training set





AdaBoost is an iterative algorithm. In the t-th iteration, each of
the 







 |
(41) |
 is the weight of the nth training samples at the t-th
iteration. We can also get the correct rate:
is the weight of the nth training samples at the t-th
iteration. We can also get the correct rate:
 |
(42) |
 , all
, all  samples are equally
weighted by
samples are equally
weighted by
 , and the weighted error
, and the weighted error
 |
(43) |
 in the training
set to be misclassified by
in the training
set to be misclassified by  . For the weak classifier the error
rate defined above only needs to be lower than 50 percent, i.e.,
. For the weak classifier the error
rate defined above only needs to be lower than 50 percent, i.e.,
 . When
. When  , this weight
, this weight  will be modified
based on whether
will be modified
based on whether  is classified correctly in the subsequent
iterations, as we will see below.
is classified correctly in the subsequent
iterations, as we will see below.
At each iteration, a strong or boosted classifier



 for the weak classifier
for the weak classifier
 in the i-th iteration is obtained as discussed
below. Taking the sign function of
in the i-th iteration is obtained as discussed
below. Taking the sign function of
 we get the
strong classifier:
we get the
strong classifier:
![$\displaystyle H_t({\bf x}_n)=sign [F_t({\bf x}_n)]=\left\{\begin{array}{ll}
+1 & F_t({\bf x}_n)>0\\ -1 & F_t({\bf x}_n)<0\end{array}\right.$](img197.svg) |
(45) |
 into one of the
two classes, while the magnitude
into one of the
two classes, while the magnitude
 represents
the confidence of the decision.
represents
the confidence of the decision.
The weight 


 |
(46) |
 of the t-th iteration can be obtained recursively
from
of the t-th iteration can be obtained recursively
from
 of the previous iteration:
of the previous iteration:
 .
.
The performance of the strong classifier can be measured by the
exponential loss, defined as the sume of all 
 |
(48) |
The coefficient 



 |
(49) |
 to zero:
to zero:
 |
(50) |
 as the optimal coefficient
that minimizes
as the optimal coefficient
that minimizes  :
:
 |
(51) |
 ,
,
 , and
, and
 |
(52) |
We now see that if a weak classifier 










Replacing 

 |
(53) |
 , i.e.,
, i.e.,  is classified
correctly by
is classified
correctly by  , it will be weighted more lightly by
, it will be weighted more lightly by
 in the next iteration, but if
in the next iteration, but if
 , i.e.,
, i.e.,  is classified incorrectly by
is classified incorrectly by  , it will be weighted more heavily by
, it will be weighted more heavily by
 in the next iteration, thereby it will be emphasized
and have a better chance to be corrected to have more accurately
classified by
in the next iteration, thereby it will be emphasized
and have a better chance to be corrected to have more accurately
classified by  in the next iteration.
in the next iteration.
We further consider the ratio of the exponential losses of two
consecutive iterations:
 |
 |
 |
|
 |
 |
||
 |
 |
(54) |
 of
of
 and
and
 , which reaches its maximum when
, which reaches its maximum when
 . However, as
. However, as
 ,
the ratio is always smaller than 1. We see that the exponential cost
can be approximated as an exponentially decaying function from its
initial value
,
the ratio is always smaller than 1. We see that the exponential cost
can be approximated as an exponentially decaying function from its
initial value
 for some
for some
 :
:
 |
(55) |
The weak classifier used in each iteration is typically implemented
as a decision stump, a binary classifier that partitions the
N-D feature space into two regions. Specifically, as a simple example,
a coordinate descent method can be used by which all training samples
are projected onto the ith dimension of the feature space 
 |
(56) |
 along the
direction of
along the
direction of  :
:
 |
(57) |
 along that direction of
along that direction of
 :
:
 |
(58) |
Alternatively, the weak classifier can also be obtained based on the principal component analysis (PCA) by partitioning the N-D feature space along the directions of the eigenvectors of the between-class scatter matrix
![$\displaystyle {\bf S}_b=\frac{1}{N}\left[N_-({\bf m}_{-1}-{\bf m})({\bf m}_{-1}-{\bf m})^T
+N_+({\bf m}_{+1}-{\bf m})({\bf m}_{+1}-{\bf m})^T\right]$](img241.svg) |
(59) |
 and
and  are the numbers of samples in the two classes
are the numbers of samples in the two classes
 , and
, and
 |
(60) |
 samples in the training set.
The training samples are likely to be better separated along these
directions of the eigenvectors of
samples in the training set.
The training samples are likely to be better separated along these
directions of the eigenvectors of  , which measures the
separability of the two classes. The eigenequations of
, which measures the
separability of the two classes. The eigenequations of  is:
is:
 |
(61) |
 is symmetric, the eigenvector matrix
is symmetric, the eigenvector matrix  is orthonormal and its columns can be used as an orthogonal basis
that spans the feature space as well as the standard basis. The
same binary threshold classification considered above for the weak
classifiers can be readily applied along these PCA bases.
is orthonormal and its columns can be used as an orthogonal basis
that spans the feature space as well as the standard basis. The
same binary threshold classification considered above for the weak
classifiers can be readily applied along these PCA bases.
The Matlab code of the main iteration loop of the algorithm is
listed below, followed by the essential functions called by the
main loop. Here T is the maximum number of iteration, and
N is the total number of training samples.
The algorithm can be carried out in the feature space based on
either the PCA basis (columns of the eigenvector matrix 

The decision stump is implemented by a binary classifier,
which partitions all training samples projected onto a 1-D space
into two groups by a threshold value, which needs to be optimal in
terms of the number of misclassification. A parameter plt is
used to indicate the polarity of the binary classification, i.e.,
which of the two class 

Plt=[]; % polarity of binary classification
Tr=[]; % transformation of basis vectors
Th=[]; % threshold of binary classification
Alpha=[]; % alpha values
h=zeros(T,N); % h functions
F=zeros(T,N); % F functions
w=ones(T,N); % weights for N training samples
Er=N; % error initialized to N
t=0; % iteration index
while t<T & Er>0 % the main iteration
t=t+1;
[tr th plt er]=WeakClassifier(X,y,w(t,:),PCA); % weak classifier
alpha=log(sqrt((1-er)/er)); % update alpha
Alpha=cat(1,Alpha,alpha); % record alpha
Tr=cat(2,Tr,tr'); % record transform vector
Th=cat(1,Th,th); % record threshold
Plt=cat(1,Plt,plt); % record polarity
x=tr*X; % carry out transform
c=sqrt(er/(1-er));
for n=1:N % update weights
h(t,n)=h_function(x(n),th,plt); % find h function
if h(t,n)*y(n)<0
w(t+1,n)=w(t,n)/c; % update weights
else
w(t+1,n)=w(t,n)*c;
end
end
F(t,:)=Alpha(t)*h(t,:); % get F functions
if t>1
F(t,:)=F(t,:)+F(t-1,:);
end
Er=sum(sign(F(t,:).*y)==-1); % error of strong classifier, number of misclassifications
fprintf('%d: %d/%d=%f\n',t,Er,N,Er/N)
end
Here are the functions called by the main iteration loop above:
function [Tr,Th Plt Er]=WeakClassifier(X,y,w,pca)
% X: N columns each for one of the N training samples
% y: labeling of X
% w: weights for N training samples
% pca: use PCA dimension if pca~=0
% Er: minimum error among all D dimensions
% Tr: transform vector (standard or PCA basis)
% Th: threshold value
% Plt: polarity
[D N]=size(X);
n0=sum(y>0); % number of samples in class C+
n1=sum(y<0); % number of samples in class C-
if pca % find PCA basis
for i=1:D
Y(i,:)=w.*X(i,:);
end
X0=Y(:,find(y>0)); % all samples in class C+
X1=Y(:,find(y<0)); % all samples in class C-
m0=mean(X0')'; % mean of C+
m1=mean(X1')'; % mean of C-
mu=(n0*m0+n1*m1)/N; % over all mean
Sb=(n0*(m0-mu)*(m0-mu)'+n1*(m1-mu)*(m1-mu)')/N; % between-class scatter matrix
[v d]=eig(Sb); % eigenvector and eigenvalue matrices of Sb
else
v=eye(N); % standard basis
end
Er=9e9;
for i=1:D % for all D dimensions
tr=v(:,i)'; % get transform vector from identity or PCA matrix
x=tr*X; % rotate the vector
[th plt er]=BinaryClassifier(x,y,w); % binary classify N samples in 1-D
er=0;
for n=1:N
h(n)=h_function(x(n),th,plt); % h-function of nth sample
if h(n)*y(n)<0 % if misclassified
er=er+w(n); % add error
end
end
er=er/sum(w); % total error of dimension d
if Er>er % record info corresponding to min error
Er=er; % min error
Plt=plt; % polarity
Th=th; % threshold
Tr=tr; % transform vector
end
end
end
function h=h_function(x, th, plt)
if xor(x>th, plt)
h=1;
else
h=-1;
end
end
function [Th Plt Er]=BinaryClassifier(x,y,w)
N=length(x);
[x1 i]=sort(x); % sort 1-D data x
y1=y(i); % reorder the targets
w1=w(i); % reorder the weights
Er=9e9;
for n=1:N-1 % for N-1 ways of binary classification
e0=sum(w1(find(y1(1:n)==1)))+sum(w1(n+find(y1(n+1:N)==-1)));
e1=sum(w1(find(y1(1:n)~=1)))+sum(w1(n+find(y1(n+1:N)~=-1)));
if e1 > e0 % polarity: left -1, right +1
plt=0; er=e0;
else % polarity: left +1, right -1
plt=1; er=e1;
end
if Er > er % update minimum error for kth dimension
Er=er; % minimum error
Plt=plt; % polarity
Th=(x1(k)+x1(k+1))/2; % threshold
end
end
end
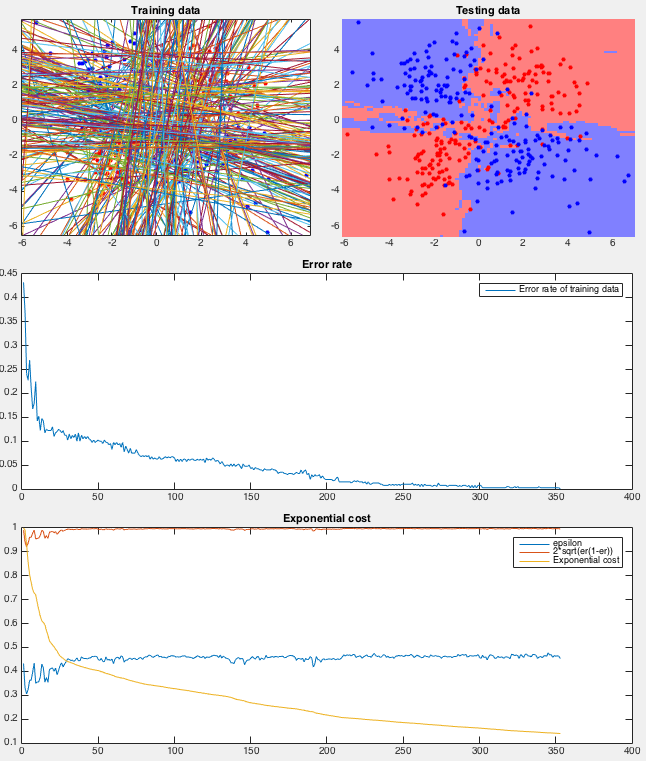
Example 0 This example shows the classification of an XOR data
set (used previously the test the naive Bayes mehtod), containing two
classes of 
After over 350 iterations the error rate eventually reduced to zero (guaranteed by the AdaBoost method). The sequence of binary participation Although all training samples are correctly classified eventually, the result suffers the problem of overfitting.
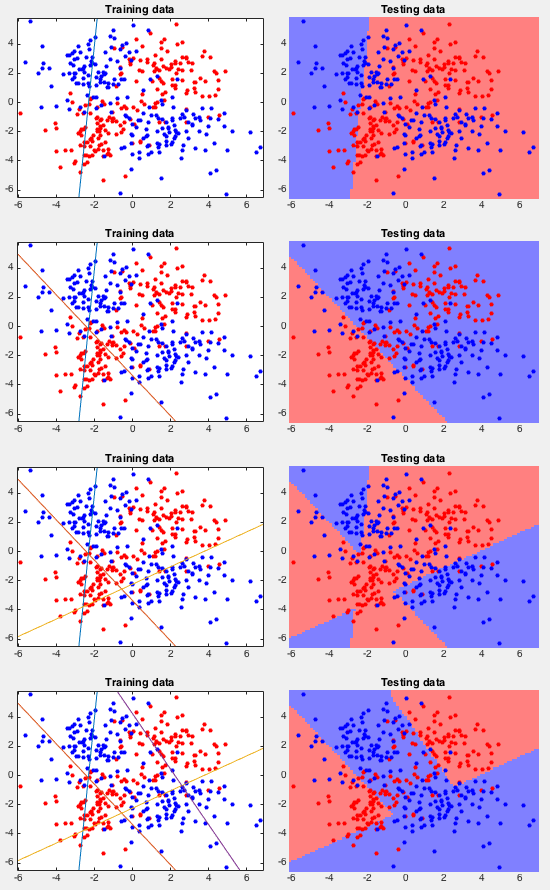
Example 1 This example shows the classification of two classes
of 
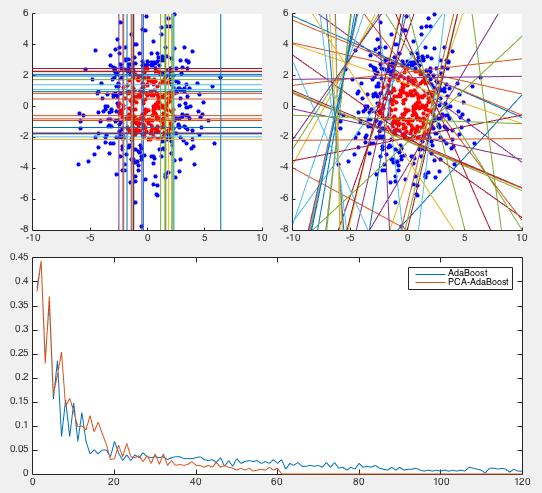
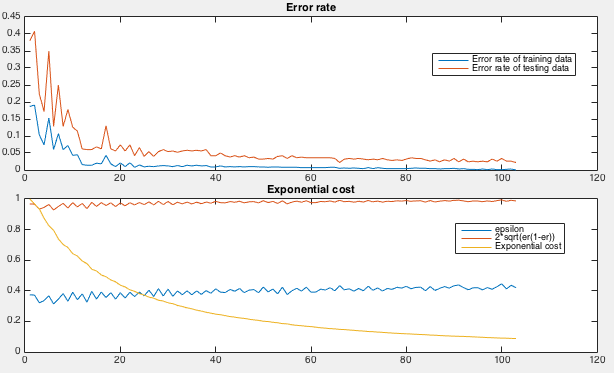
How the AdaBoost is iteratively trained can be seen in the figure below.
First, the error rate, the ratio between the number of misclassified
samples and the total number of samples, for both the training and
testing samples, are plotted as the iteration progresses (top). Also,
the weighted error






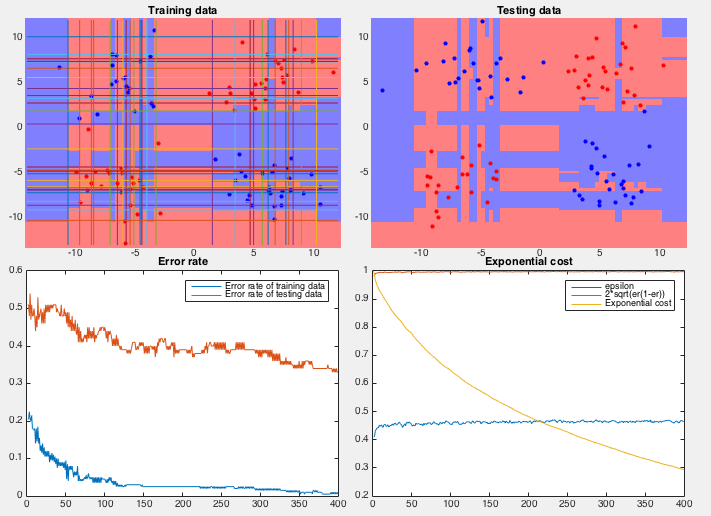
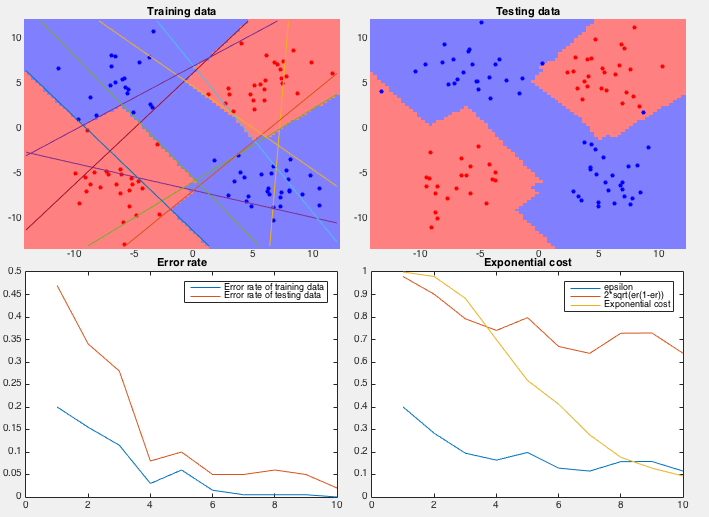
Example 2 In this dataset, 100 training samples of two classes (50 each) forms four clusters arranged in the 2-D space as an XOR pattern as shown figure (top-left), and 100 testing samples of the same distribution are classified by the AbaBoost algorithm trained by both the coordinate descent and PCA methods.
We see that the coordinate descent method performs very poorly in both the slow convergence (more than 400 iterations) during training and high classification error rate (more than 1/3) during testing. The partitioning of the 2-D space is an obvious over fitting of the training set instead of reflecting the actual distribution of the two classes (four clusters). On the other hand, the PCA method converges quickly (10 iterations) and classifies the testing samples with very low error rate. The space is clearly partitioned into two regions corresponding to the two classes.