Next: Karhunen-Loeve Transformation Up: Principal Component Analysis Previous: Principal Component Analysis
In principal component analysis (PCA), each pattern 

 |
 |
![$\displaystyle E[ {\bf x}_i ]=\int x_i\, p(x_i)\, dx_i$](img150.svg) |
|
 |
 |
![$\displaystyle E[ (x_i-\mu_i)^2 ]=E[x_i^2]-\mu_i^2
=\int x_i^2\,p(x_i)\, dx_i-\mu_{x_i}^2,\;\;\;\;
(i=1,\cdots,d)$](img152.svg) |
(37) |
 and
and  is:
is:
 |
 |
![$\displaystyle E[ (x_i-\mu_i)(x_j-\mu_j) ]=E[ x_ix_j ]-\mu_i\mu_j$](img155.svg) |
|
 |
 |
(38) |
 are:
are:
 |
 |
![$\displaystyle E[ {\bf x} ]=\left[\begin{array}{c}
\mu_1\\ \vdots\\ \mu_d\end{array}\right]$](img158.svg) |
|
 |
 |
![$\displaystyle E[ ({\bf x}-{\bf m}_x)({\bf x}-{\bf m}_x)^T ]
=E[ {\bf xx}^T ]-{\...
... & \ddots & \vdots \\
\sigma_{d1}^2 & \cdots & \sigma_{dd}^2\end{array}\right]$](img160.svg) |
(39) |
Usually the joint probability density function





![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img6.svg)
Note that the rank of the





 |
(41) |
The variance
![$\sigma_i^2=E[(x_i-\mu_{x_i})^2]$](img170.svg)




![$\sigma_{ij}^2=E[(x_i-\mu_{x_i})(x_j-\mu_{x_j})]$](img172.svg)





 |
(42) |
 and
and  are
correlated.
are
correlated.
 or
or
 :
:  and
and  are maximally
correlated. The information contained in the two variables is
completely redendant, given the value of one of them, the value
of the other is known.
are maximally
correlated. The information contained in the two variables is
completely redendant, given the value of one of them, the value
of the other is known.
 or
or
 :
:  and
and  are correlated
to different extents. The information they each carry has certain
redendancy.
are correlated
to different extents. The information they each carry has certain
redendancy.
 :
:  and
and  are uncorrelated. They each
carry their own independent information with no redencdancy.
are uncorrelated. They each
carry their own independent information with no redencdancy.
 that measures how
much two random variables are correlated also measures the redendancy
of the information they carry. When there exists some data redendancy
in the data, it is possible to carry out data compression by some
method such as the principal component analysis based on the covariance
that measures how
much two random variables are correlated also measures the redendancy
of the information they carry. When there exists some data redendancy
in the data, it is possible to carry out data compression by some
method such as the principal component analysis based on the covariance
 to reduce the data redendancy, so that the data size
can be significantly reduced while the information (dynamic energy)
contained in the data is still mostly preserved.
to reduce the data redendancy, so that the data size
can be significantly reduced while the information (dynamic energy)
contained in the data is still mostly preserved.
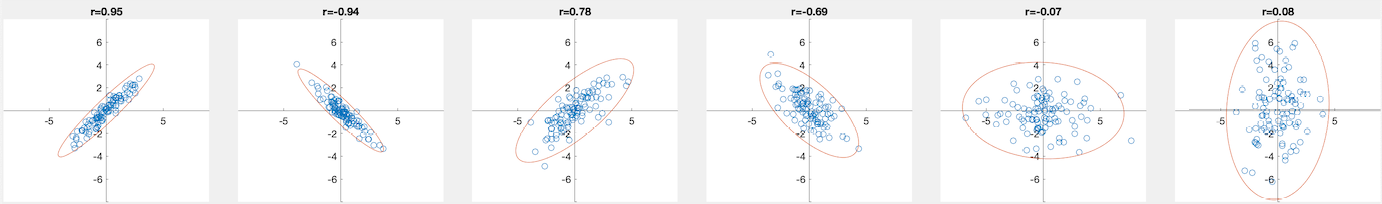
Examples
Six normally distributed 2-D datasets are generated with zero mean and the following covariance matrices:
![$\displaystyle {\bf\Sigma}_1=\left[\begin{array}{rr}
1.0 & 0.95 \\ 0.95 & 1\end{...
...\Sigma}_3=\left[\begin{array}{rr}
1 & 0 \\ 0 & 3\end{array}\right],\;\;\;\;\;\;$](img182.svg) |
(43) |
These data points are plotted below, together with the correlation coefficient on top of each dataset.