Next: Factor Analysis and Expectation Up: ch8 Previous: Kernel Methods
In kernel PCA, the PCA method, which maps the dataset to a low dimensional space, and the kernel method, which maps the datad set to a high dimensional space, are combined to take advantage of both the high dimensional space (e.g., better separability) and the low dimensional space (e.g., computational efficiency).
Specifically, we assume the given dataset
![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img6.svg)
![${\bf Z}=[{\bf z}_1,\cdots,{\bf z}_N]$](img413.svg)

 |
(108) |
 data points in the dataset
data points in the dataset  , we can further define
a kernel matrix:
, we can further define
a kernel matrix:
![$\displaystyle {\bf K}={\bf Z}^T{\bf Z}
=\left[\begin{array}{c}{\bf z}_1^T\\ \vd...
...&\cdots&k_{1N}\\ \vdots&\ddots&\vdots\\
k_{N1}&\cdots&k_{NN}\end{array}\right]$](img415.svg) |
(109) |
To carry out KLT in the high dimensional space, we need to estimate
the covariance matrix of

 |
(110) |
![${\bf Z}=[{\bf z}_1,\cdots,{\bf z}_N]$](img413.svg) have a zero mean
have a zero mean
 , which will be justified later,
and get (same as Eq. (82)):
, which will be justified later,
and get (same as Eq. (82)):
![$\displaystyle {\bf\Sigma}_z
=\frac{1}{N}\sum_{n=1}^N ({\bf z}_n-{\bf m}_z)({\bf...
...{c}
{\bf z}_1^T\\ \vdots\\ {\bf z}_N^T\end{array}\right]
=\frac{1}{N}{\bf ZZ}^T$](img418.svg) |
(111) |
 on both sides of the eigenequation
above to get (same as Eq. (83)):
where
on both sides of the eigenequation
above to get (same as Eq. (83)):
where
 . Solving this eigenequation of
the
. Solving this eigenequation of
the  matrix
matrix
 , we get its
eigenvectors
, we get its
eigenvectors
 ,
corresponding to the eigenvalues
,
corresponding to the eigenvalues
 ,
which are the same as the non-zero eigenvalues of
,
which are the same as the non-zero eigenvalues of
 . The eigenvectors
. The eigenvectors  of
of
 in Eq. (112) can now be written
as:
in Eq. (112) can now be written
as:
 |
(114) |
 :
:
 |
 |
 |
|
 |
 |
(115) |
 of
of  are rescaled to
satisfy
are rescaled to
satisfy
 , then the eigenvectors
, then the eigenvectors  of
of
 are normalized to satisfy
are normalized to satisfy
 . Now we
can carry out PCA in the high dimensional space by
. Now we
can carry out PCA in the high dimensional space by
![$\displaystyle {\bf y}=[{\bf v}_1,\cdots,{\bf v}_N]^T{\bf z}={\bf V}^T{\bf z}$](img437.svg) |
(116) |
 is the projection of
is the projection of
 onto one of the eigenvectors
onto one of the eigenvectors  of
of
 :
:
 and
and
![${\bf k}=[k_1,\cdots,k_N]^T$](img443.svg) . Although neither
. Although neither  nor
nor
 is available, their inner product
is available, their inner product
 can still be obtained based on the kernel
function
can still be obtained based on the kernel
function
 . We see that all data points in
the high dimensional space appear only in the form of an inner
product in the equation above, as well as in the eigenequation
Eq. (113), the kernel mapping
. We see that all data points in
the high dimensional space appear only in the form of an inner
product in the equation above, as well as in the eigenequation
Eq. (113), the kernel mapping
 never needs to be explicitly carried out.
never needs to be explicitly carried out.
The discussion above is based on the assumption that the data
in the high dimensional space have zero mean





 |
 |
 |
|
 |
 |
||
 |
 |
(118) |
 such inner products
can be written as:
where
such inner products
can be written as:
where
![$\displaystyle {\bf 1}_N=\frac{1}{N}\left[\begin{array}{ccc}
1&\cdots&1\\ \vdots&\ddots&\vdots\\ 1&\cdots&1\end{array}\right]_{N\times N}$](img453.svg) |
(120) |
If we replace the kernel matrix 





 , obtain
, obtain
 based on kernel mapping
based on kernel mapping
 for
all
for
all
 , and then find
, and then find
 , as in Eq.
(119);
, as in Eq.
(119);
 to get the eigenvalue
to get the eigenvalue
 and the corresponding eigenvector
and the corresponding eigenvector  for all
for all
 ;
;
 (implicit)
onto the nth eigenvector
(implicit)
onto the nth eigenvector  to get the principal component:
to get the principal component:
 for each
for each
 , as in Eq. (117).
, as in Eq. (117).
 principal components
principal components
 in the high dimensional feature space, containing
most of the information.
in the high dimensional feature space, containing
most of the information.
Note that in kernel PCA methods, the complexity for solving the
eigenvalue problem of the 





Shown below is a Matlab function for the kernel PCA algorithm
which takes the dataset
![${\bf X}=[{\bf x}_1,\cdots,{\bf x}_N]$](img6.svg)


Kernel(X,X) calculates the kernel matrix 
function [Y d]=KPCA(X)
[D N]=size(X);
K=Kernel(X,X); % kernel matrix of data X
one=ones(N)/N;
Kt=K-one*K-K*one+one*K*one; % Kernel matrix of data with zero mean
[U D]=eig(Kt); % solve eigenvalue problem for K
[d idx]=sort(diag(D),'descend'); % sort eigenvalues in descending order
U=U(:,idx);
D=D(:,idx);
for n=1:N
U(:,n)=U(:,n)/d(n)/N;
end
Y=(K*U)';
end
Examples:
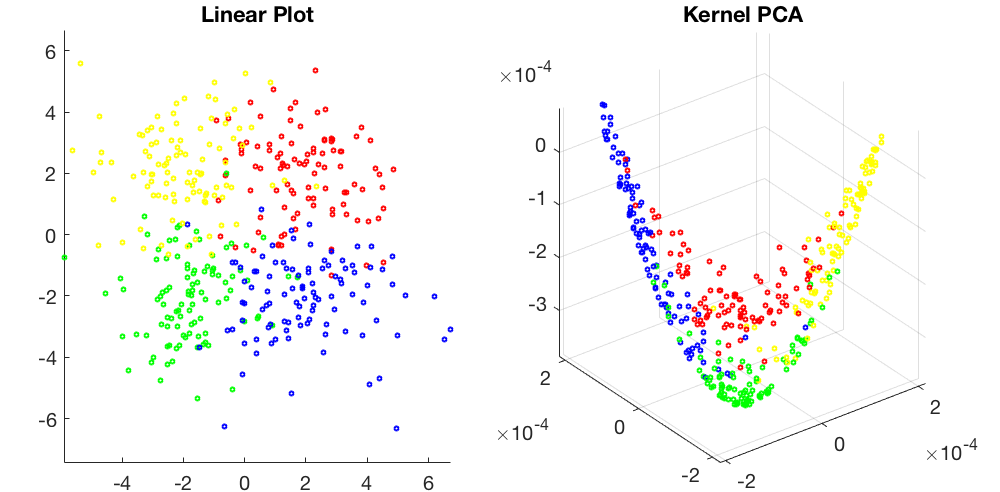
Visualization of a 2-D dataset of four classes, linear plot on the left and kernel PCA (based on RBF kernel) on the right:
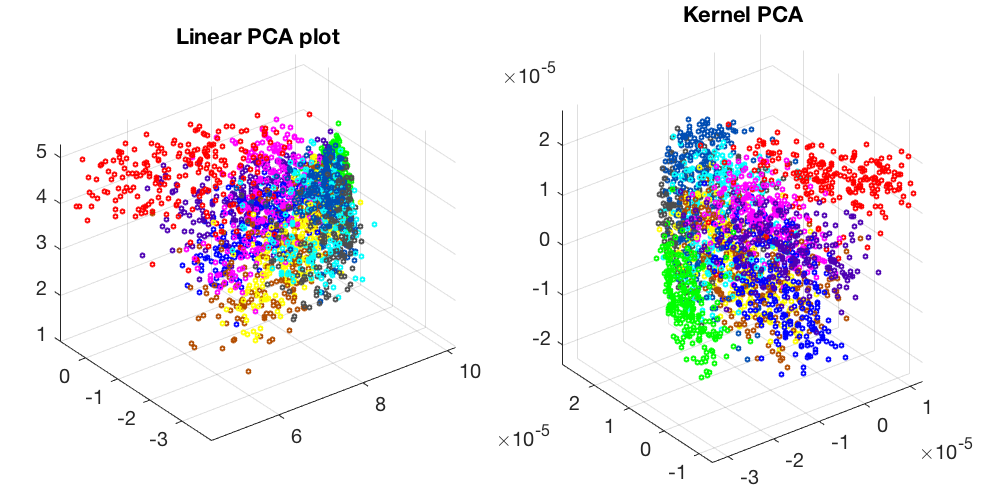
Visualization of a 256-D dataset of 10 classes of hand-written digits:
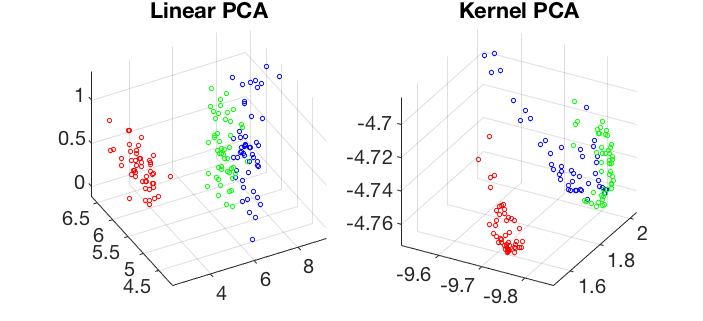
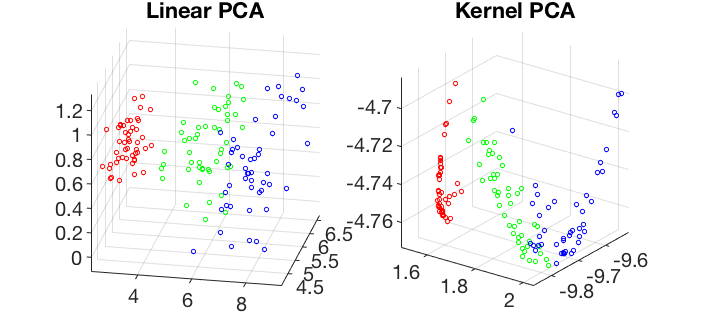
Visualization of the 4-D iris dataset in 3-D space spanned by the first 3 principal components of the linear and kernel PCA transforms: